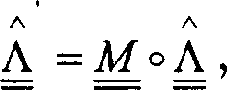Inferring gene regulatory networks from time-ordered gene expression data using differential equations
A technique of differential equations and gene expression, which is applied in the field of determining the relationship between genes in organisms, and can solve problems such as inaccessibility
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
example 1
[0087] Example 1: Gene network in Bacillus subtilis
[0088] Embodiments of the invention for finding gene regulatory networks using gene expression data were recently measured in MMGE gene expression experiments in Bacillus subtilis. MMGE is a synthetic minimal medium containing glucose and glutamine as carbon and nitrogen sources. In this medium, the expression of genes required for the biosynthesis of small molecules, such as amino acids, is induced. In this experiment, the expression level of the 4320 ORF was measured at eight time points in one hour intervals, with two measurements at each time point.
[0089] Data Preparation and Analysis
[0090] To reduce the effect of measurement noise present in the data, the expression level of each gene was compared to the measured background level. Genes with an average gene expression level below the average background level, whether in the red or green channel, were removed from the analysis.
[0091] Global normalization wa...
example 2
[0099] Example 2: Clustering of Bacillus subtilis genes
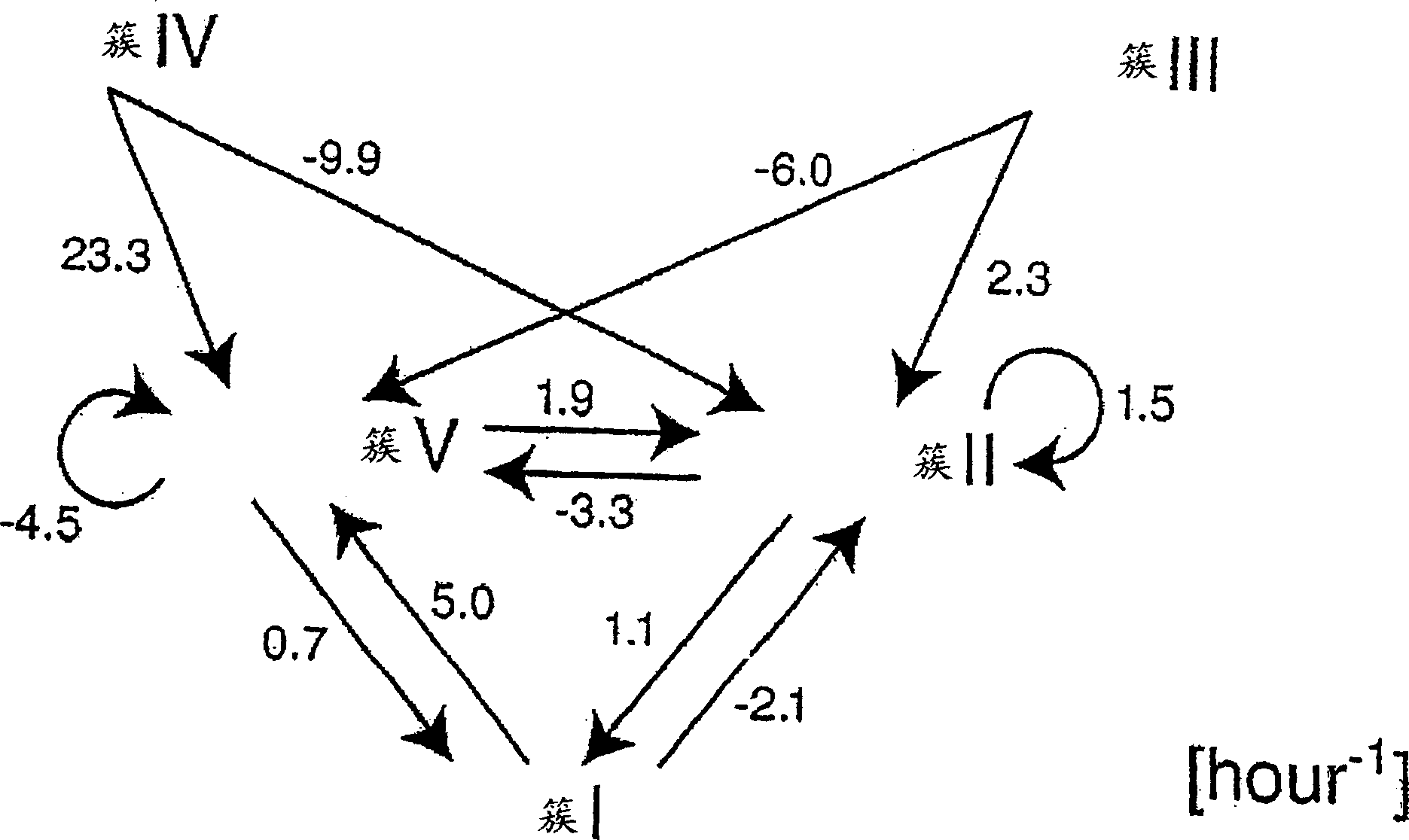
[0100] These 684 genes of B. subtilis were clustered sequentially into 5 groups using k-means clustering. The distance between genes was measured using Euclidean distance, and the centroid of a cluster was defined as the median of all genes in the cluster. The number of clusters was chosen to avoid significant overlap. The k-means algorithm is repeated 1,000,000 times starting with different random initial clusters. Find the best solution 81 times.
[0101] Complete clustering results are available at the following websites:
[0102] http: / / bonsai.ims.u-tokyo.ac.jp / -mdehoon / publications / Subtilis / clusters.html.
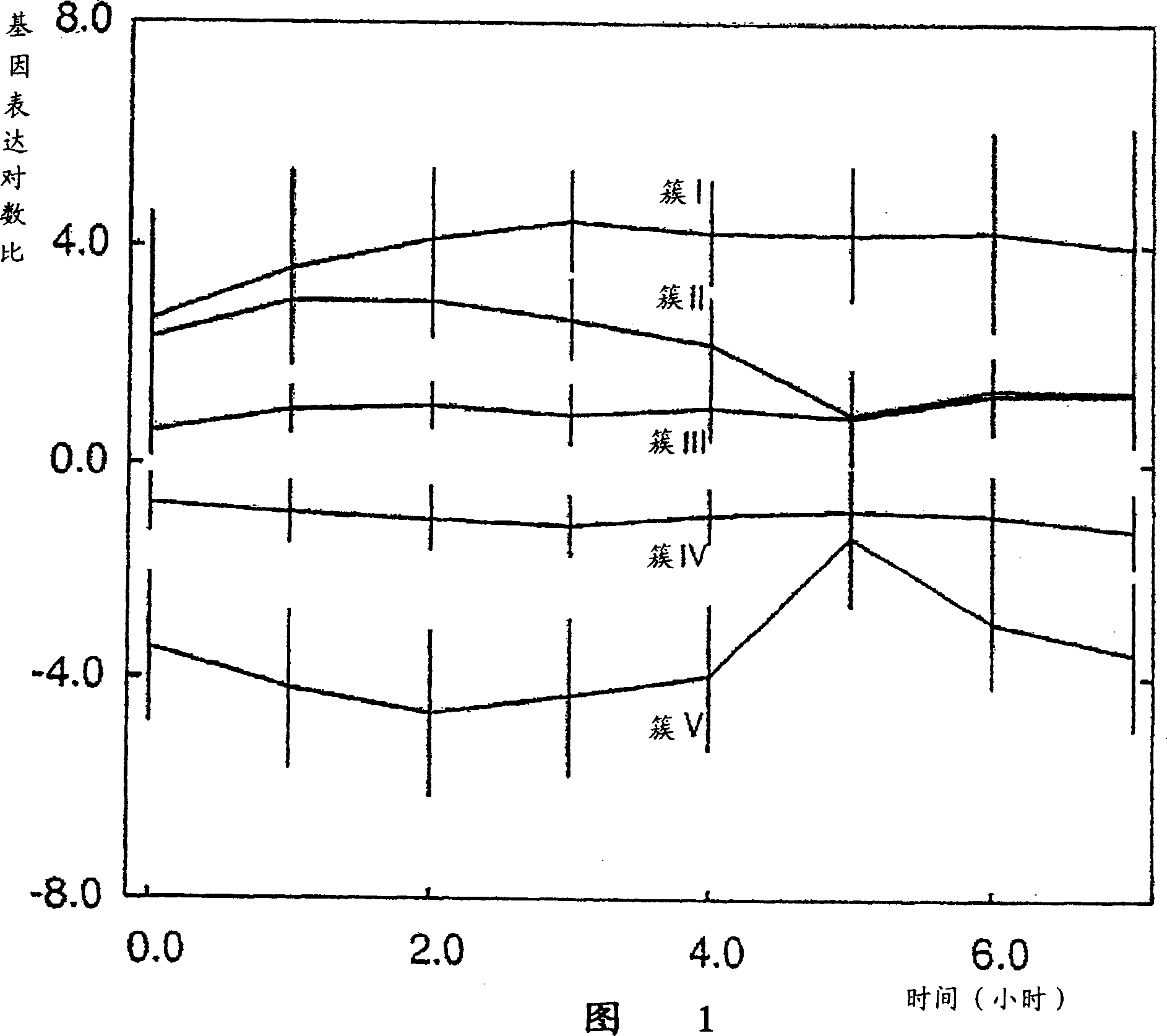
[0103] To determine the biological function of the created clusters, we considered the functional categories of all genes in each cluster in the Bacillus subtilis database. Table 1 lists the main functional categories of the five clusters formed.
[0104] Figure 1 represents the log ratio of gene expr...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com