Establishment method of gene regulating network based on two-way XGBoost
A technology for gene regulation network and construction method, applied in special data processing applications, instruments, electrical digital data processing, etc.
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment Construction
[0050] The present invention will be further described in detail below in conjunction with the accompanying drawings and specific embodiments,
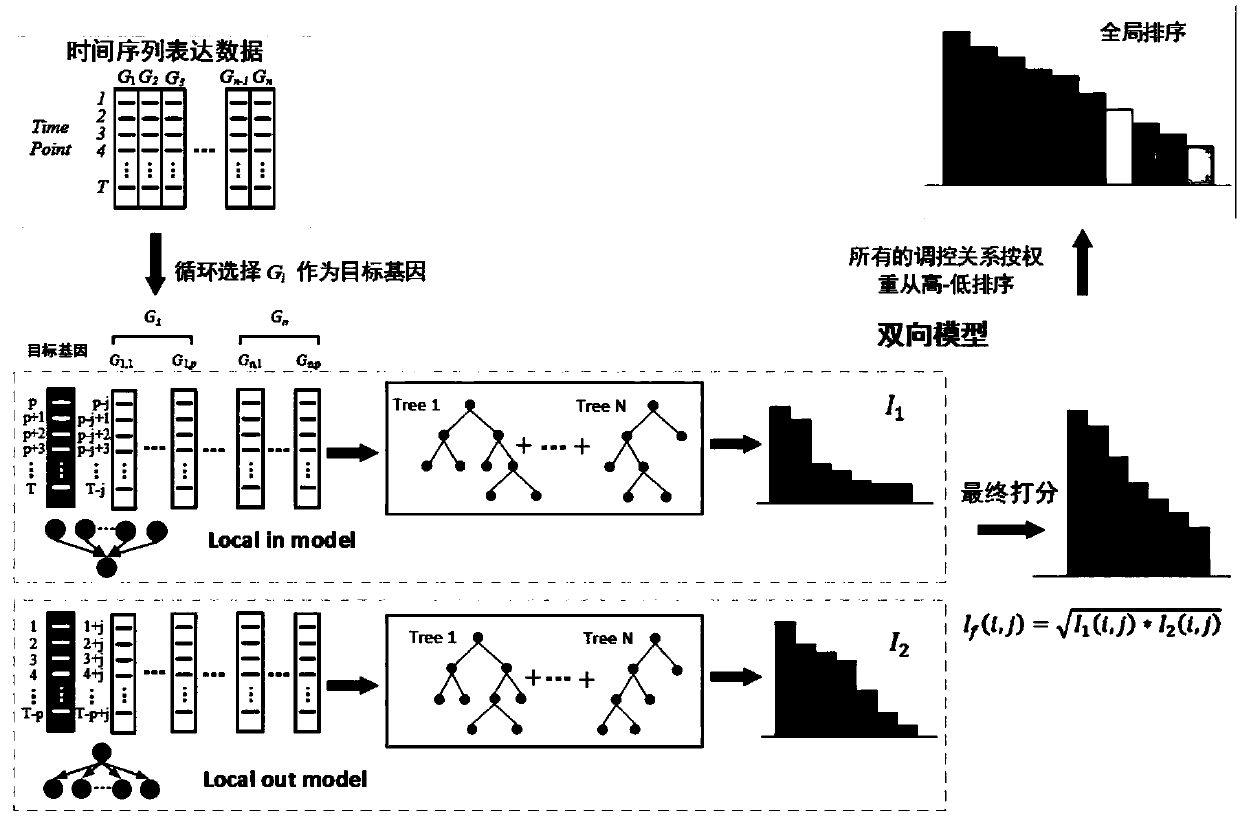
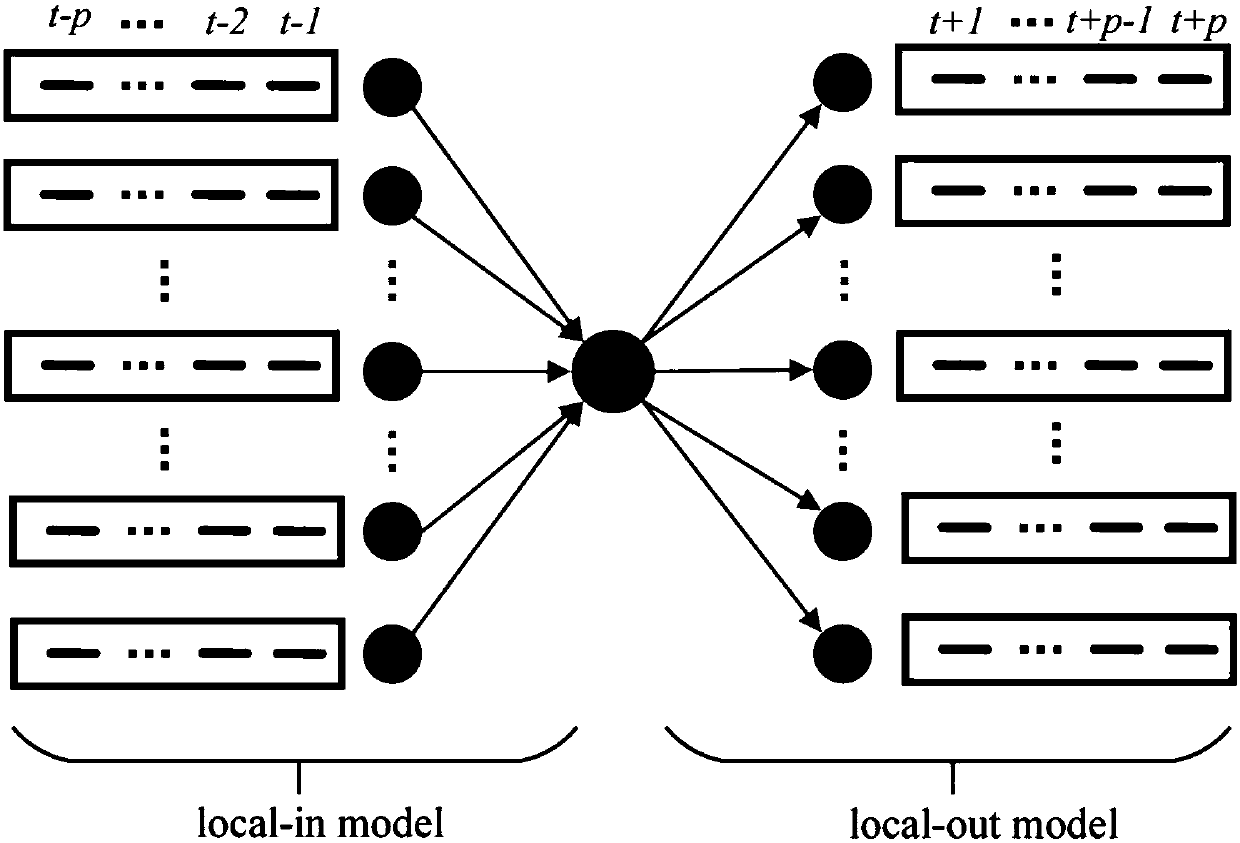
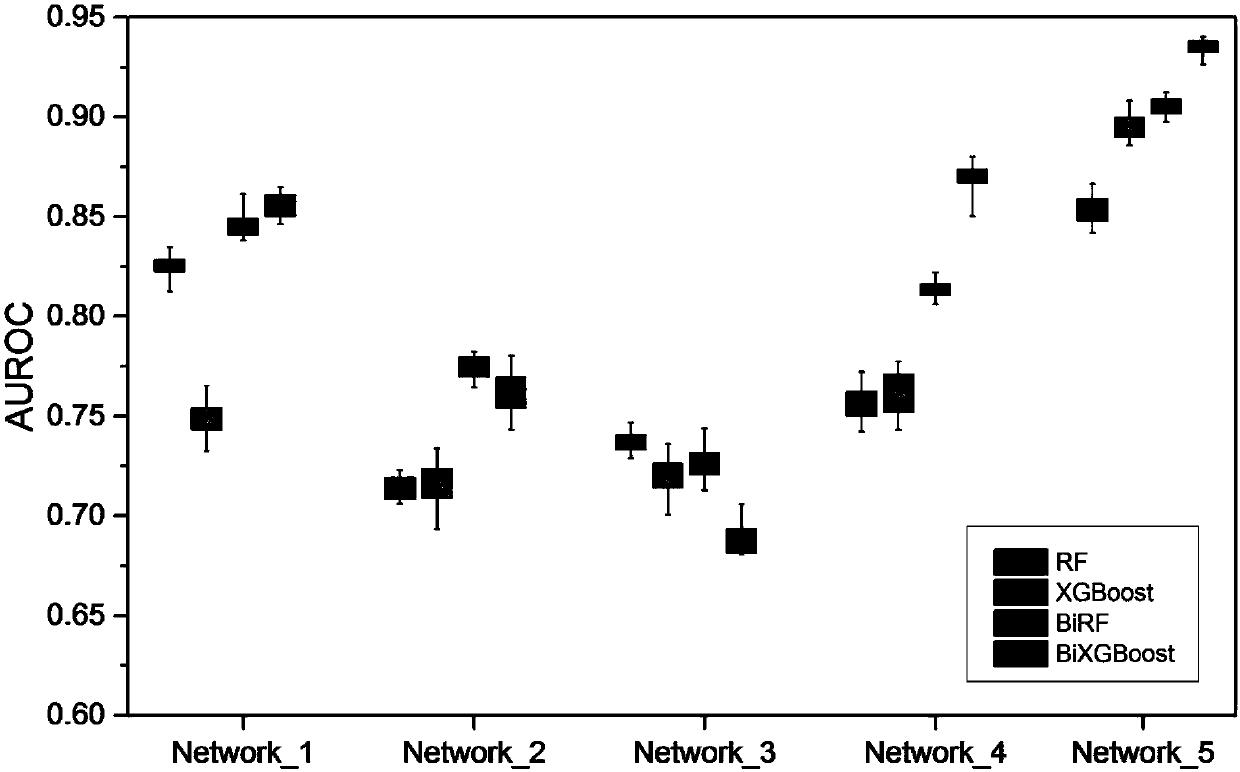
[0051] The invention discloses a method for constructing a gene regulation network based on bidirectional XGBoost. According to the characteristics of time series gene expression data, the relationship between the expression value of a gene at time t and other genes before and after time t is analyzed, and local-in and local -out two-way model; in a single model, the construction of the gene regulatory network is divided into multiple regression equations, fully considering the situation of high-order time lag, using XGBoost to evaluate the weight of the features in a single regression equation, and finally obtain the ranking of the regulatory relationship ; Use the normalized residual to reweight the features in each independent regression equation twice, and integrate the weight of the two-way model on the same pair of regulatory rel...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com