Gene module analysis method
An analysis method and gene technology, applied in the field of data analysis, can solve the problems of gene module deviation, lack of high dimension, and inability to obtain disease module structure.
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment Construction
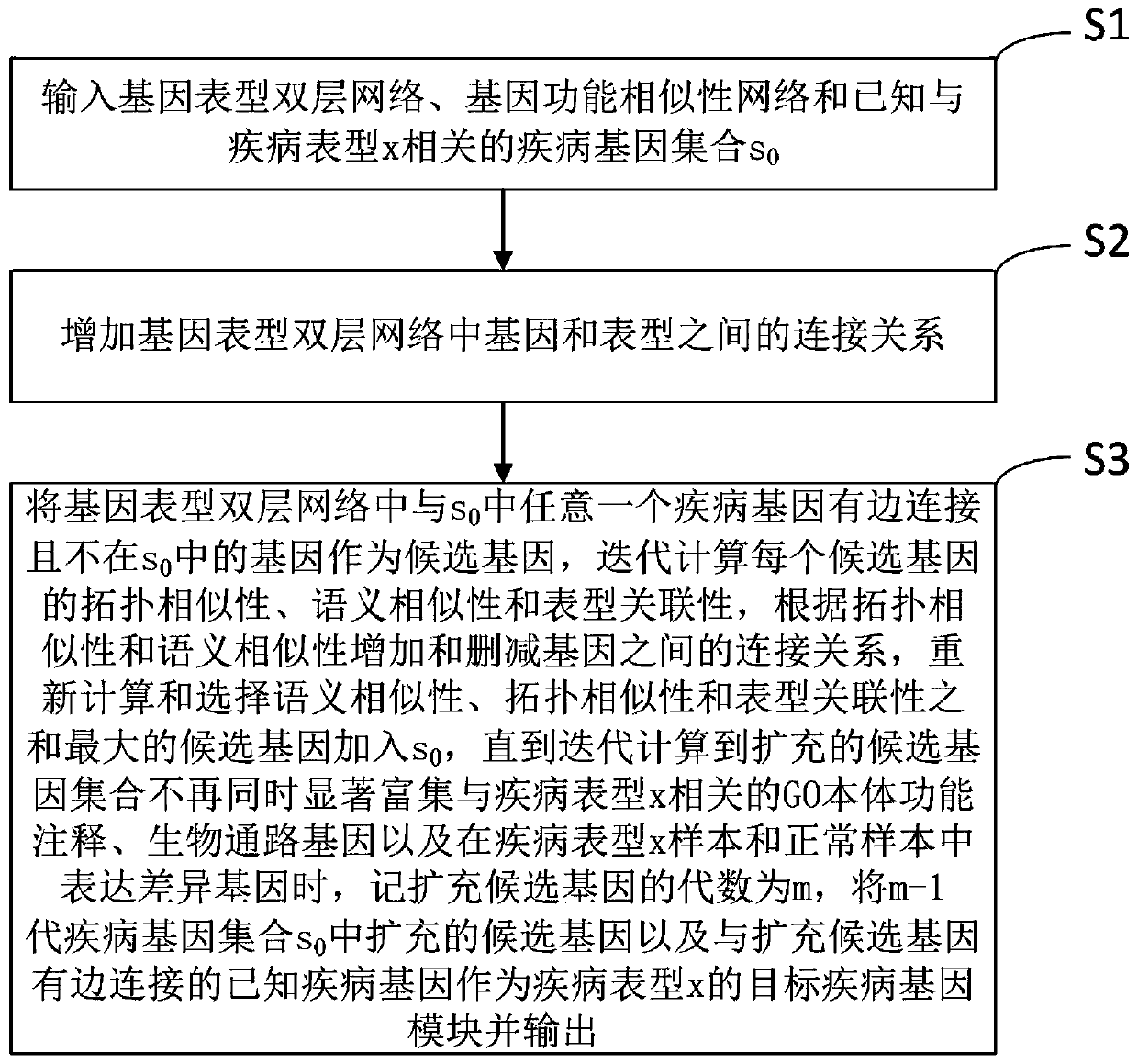
[0038] refer to figure 1 , a kind of gene module analysis method that the present invention proposes, comprises:
[0039] S1, input gene-phenotype double-layer network, gene function similarity network and disease gene set s known to be related to disease phenotype x 0 ;
[0040] In this step, the genotype double-layer network is specifically: construct a genotype double-layer network according to the gene network A, the phenotype similarity network B, and the gene-phenotype relationship network C, and the adjacency matrix of the double-layer network can be expressed as where C T is the transpose matrix of C.
[0041] In a specific scheme, collecting D 1 l in a database 1 For protein interaction, which contains N proteins, the protein network can be abstracted as a protein point set V 1 and interacting edge set E 1 Composition of graph G 1 =(V 1 ,E 1 ), the number of nodes is denoted as N=|V 1 |, the number of sides is recorded as l 1 =|E 1 |,E 1 Each edge in ha...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com