Exogenous miRNA regulatory target gene prediction method containing three-dimensional free energy
A target gene prediction, exogenous technology, applied in the direction of instrument, sequence analysis, calculation, etc., to achieve the effect of improving accuracy, accurate feature input vector, and improving traditional sequence matching features
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
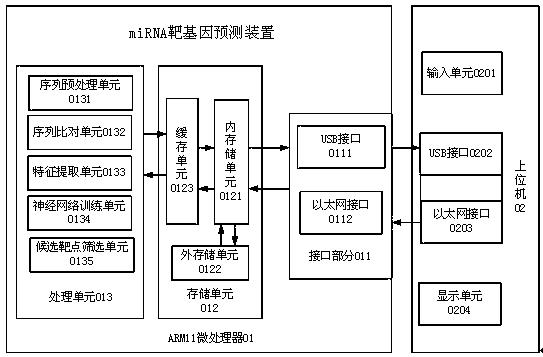
[0060] Such as figure 1 Shown, the present invention comprises ARM11 microprocessor 01, upper computer 02;
[0061] In the embodiment of the present invention, generally utilize a general-purpose PC computer as host computer 02, this host computer can be identified by the RNA secondary structure of the microprocessor based on the 32-bit S3C6410 core that Samsung produces The device is connected to work together to complete the recognition of RNA secondary structure;
[0062] Both the input unit 0201 and the display unit 0204 of the upper computer 02 use the input and output devices of the PC computer to realize their functions.
[0063] In the present invention, the intercommunication between the host computer 01 and the ARM9 microprocessor 02 can be realized through the Ethernet interface 0203 of the host computer 01 and the Ethernet interface 0204 of the ARM11 microprocessor 02, and the Ethernet interface adopts DM9000 which is fully integrated and has low cost single Fas...
Embodiment 2
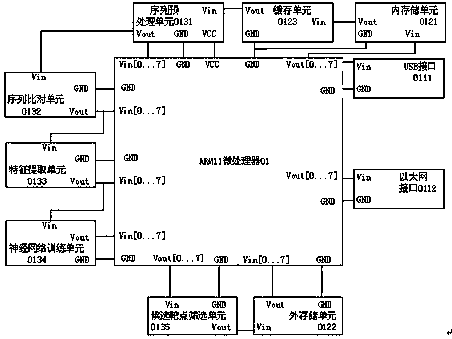
[0072] Such as image 3 Shown is the logical function diagram of the present invention. The steps for users to use the device to predict miRNA target genes are as follows:
[0073] 1) Input the miRNA sequence and target sequence through the input unit 0201 of the host computer, and transmit them to the internal storage unit 0121 through the Ethernet interface 0203, and further read the data into the cache unit 0123;
[0074] 2) The sequence preprocessing unit 0131 reads the miRNA sequence and the target sequence from the buffer unit 0123, and judges the format of the input sequence. If the data does not meet the specification, it immediately sends a high-priority error signal and transmits it through the buffer unit 0123 to The internal storage unit 0121 is then transmitted back to the display unit 0204 of the host computer through the Ethernet interface 0203 to output error information; if the sequence format is judged, a sliding window is constructed based on the length of ...
Embodiment 3
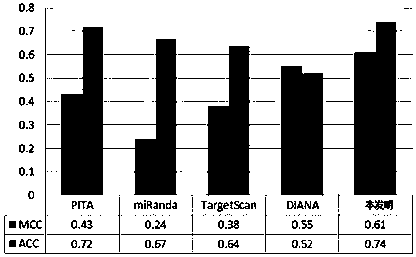
[0091] Input the miRNA sequence of hsa-let-7b through the host computer 02: TGAGGTAGTAGGTTGTGTGGTT, and then input the gene sequence of gi|332384207|gb|JF915190.1| Influenza A virus (A / California / 04 / 2009(H1N1)) PB2.
[0092] After calculation, three predicted targets are obtained:
[0093] No.: 1 score: 1.02 position: 767-783;
[0094] No.: 2 score: 1.0 position: 767-784;
[0095] No.: 3 score: 0.97 position: 767-785.
[0096] The present invention adopts classic double luciferase verification method:
[0097] 1. Select the Marc-145 cell line as the host cell, and select the psi-CHECK2 plasmid as the basis for vector construction.
[0098] 2. Amplify the DNA fragment of the GFP (green fluorescent protein) gene, the DNA fragment of the target gene and the DNA fragment of the YMP gene (yellow fluorescent protein).
[0099] 3. Carrier construction:
[0100] 1. After digestion, connect the GFP gene fragment and the target gene fragment into GFP-target gene.
[0101] 2. Dig...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com