MicroRNA (micro ribonucleic acid) related to soybean phytophthora resistance
A technology against Phytophthora root rot and Phytophthora, applied in the field of microRNA, can solve the problem of loss of resistance of disease-resistant varieties
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
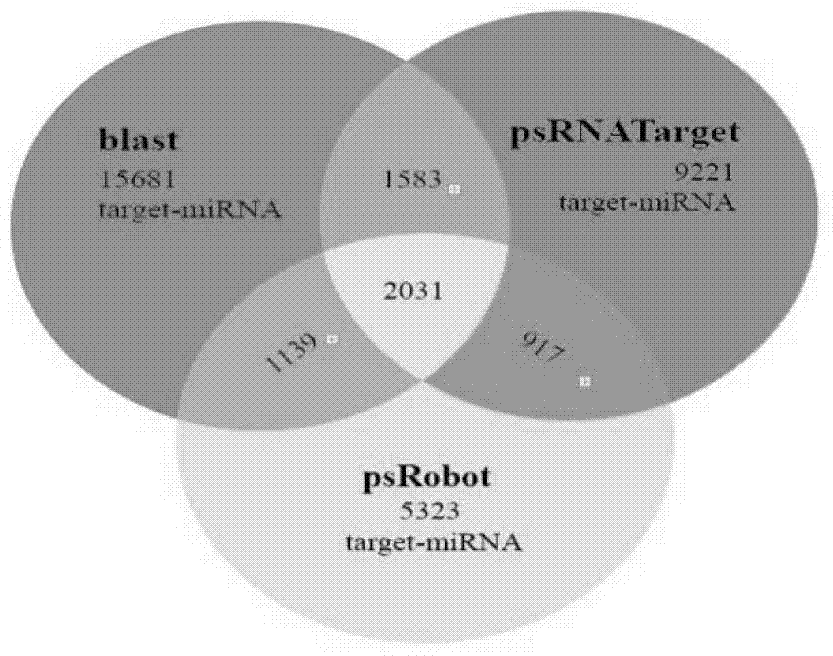
[0016] Example 1 Prediction and screening of disease-resistant gma-miR482c-3p
[0017] (1) Screening genes related to Phytophthora root rot resistance in soybean
[0018] The expression profiling of Suinong No. 10 soybean variety infected with Phytophthora root rot physiological race No. 1 was carried out. Five samples of SCKL, SML, STL0.5, STL2 and STL4 infecting the same physiological race 1 of Phytophthora root rot were used, and total RNA was extracted as sequencing samples with the RNeasy Mini Kit of Qiagen Company. Expression profile sequencing was completed by BGI. Among them, SCKL is a sample without any treatment, SML is a sample not inoculated with Phytophthora root rot, and STL0.5, STL2, and STL4 are samples taken at 0.5h, 2h, and 4h after inoculation with Phytophthora root rot. Inoculation location and method such as Figure 4 As shown, in the expression profile sequencing data of the five samples of SCKL, SML, STL0.5, STL2, and STL4, a total of 33683 spliced ...
Embodiment 2
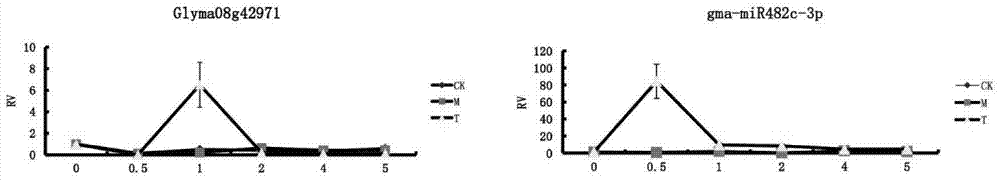
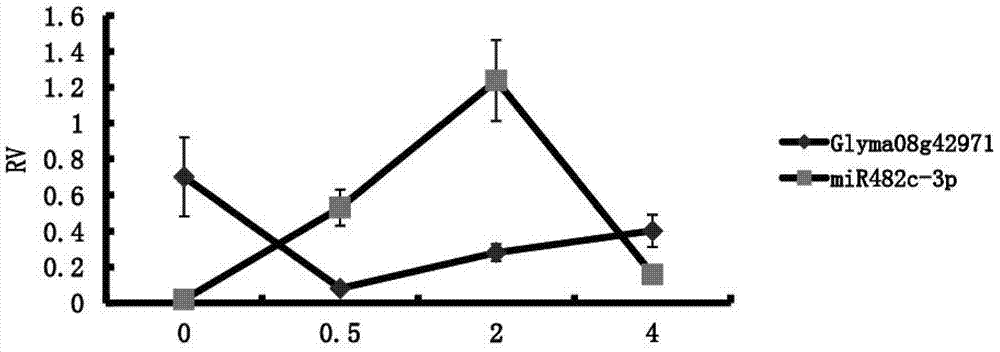
[0025] Embodiment 2 Phytophthora root rot resistance inspection
[0026] According to prediction, the target gene of gma-miR482c-3p is Glyma08g42971, which is differentially expressed during soybean infection with Phytophthora, and is related to both disease resistance and immunity in SoyBase. In this example, for gma-miR482c-3p, the gma-miR482c-3p in Suinong No. 10 soybean variety was tested for resistance to Phytophthora root rot by using the designed specific reverse transcription primer and specific PCR primer pair.
[0027] First of all, the total RNA in the sample should be extracted, and the total RNA of each sample should be reverse transcribed twice, one is to reverse transcribe gma-miR482c-3p according to the specific reverse transcription primer, and the other is to reverse transcribe gma-miR482c-3p according to the general RNA Reverse transcription primers reverse transcribe all RNA. Next, quantitative PCR was performed on the two reverse transcription products us...
Embodiment 3
[0060] Example 3 Cluster analysis of miRNA-target gene relationship
[0061] (1) Use the clustalx software to perform a pairwise comparison of the miRNAs and genes screened and predicted in Example 1, and present the pairwise comparison results using mega to obtain a guide tree. And the miRNAs and target genes were classified according to the distance between the two genes presented in the guide tree.
[0062] (2) Due to the short miRNA sequence and high family similarity, the miRNAs were divided into family MIRNA forms, and then the miRNA families and target genes were linked.
[0063] (3) Classify and organize the relationship between miRNA and target gene in the connection, as shown in Table 4, it is found that among the predicted miRNA-target gene relationships, most of the miRNAs have a tendency to regulate their target genes, according to With this tendency (Table 3), the miRNA-target gene relationship can be divided into 8 categories a1b1, a1b2, a1b3, a21b3, a22b1, a22...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com