Application of an Epstein-Barr virus miRNA associated with nasopharyngeal carcinoma
A technology for Epstein-Barr virus and nasopharyngeal carcinoma, applied in the field of molecular biology, can solve problems such as limitations in early detection, inability to distinguish whether patients are infected or cured, and non-specific reactions have a large impact
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0036] Example 1: Detection of differentially expressed miRNAs between nasopharyngeal carcinoma tissues and corresponding adjacent normal tissues by miRNA chips
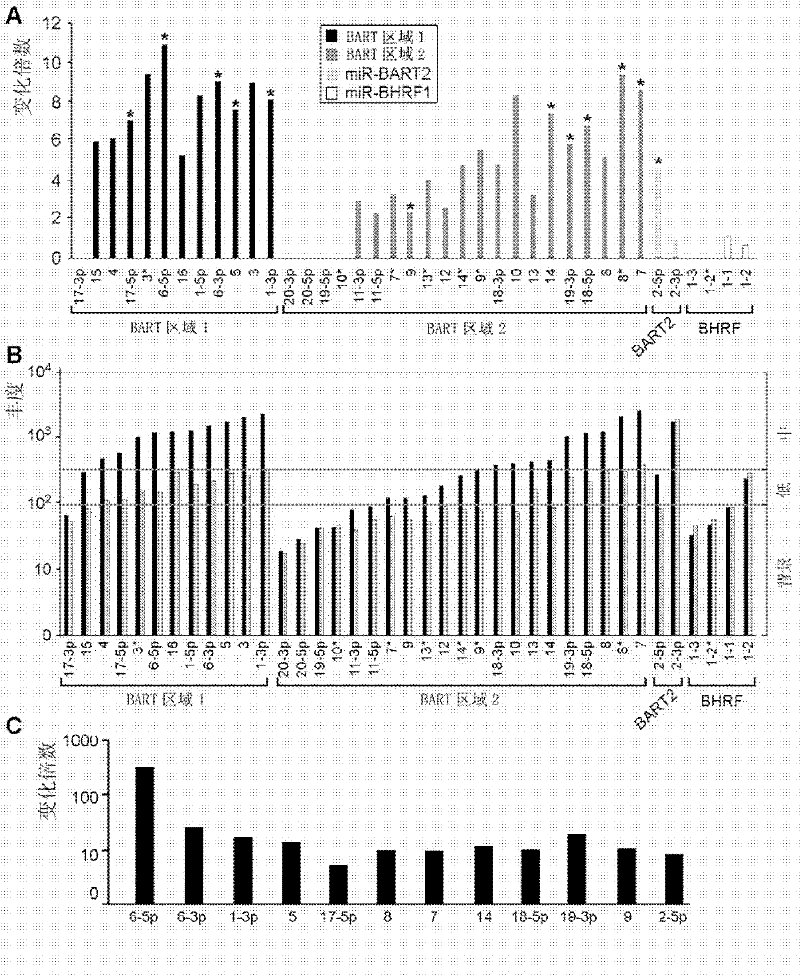
[0037] The present invention uses a micro-RNA (miRNA) chip (including 850 human and 39 EBV miRNAs, Exiqon product miRCURYLINA Array, Exiqon, Vedbaek, Denmark) to compare the miRNA expression differential profiles of five pairs of nasopharyngeal carcinoma samples. The analysis method of the chip is carried out according to the steps of the product instructions. The results showed that, compared with adjacent normal nasopharyngeal tissues, 60 miRNAs were expressed differently in nasopharyngeal carcinoma tissues, among which 50 miRNAs were upregulated and 10 miRNAs were downregulated. Among the 50 highly expressed miRNAs, 29 were encoded and transcribed by Epstein-Barr virus DNA. Table 1 lists the miRNAs with differential expression between nasopharyngeal carcinoma tissues and corresponding adjacent normal tissues dete...
Embodiment 2
[0042] Embodiment 2: A preferred embodiment of the miRNA differential expression profile experiment of EBV
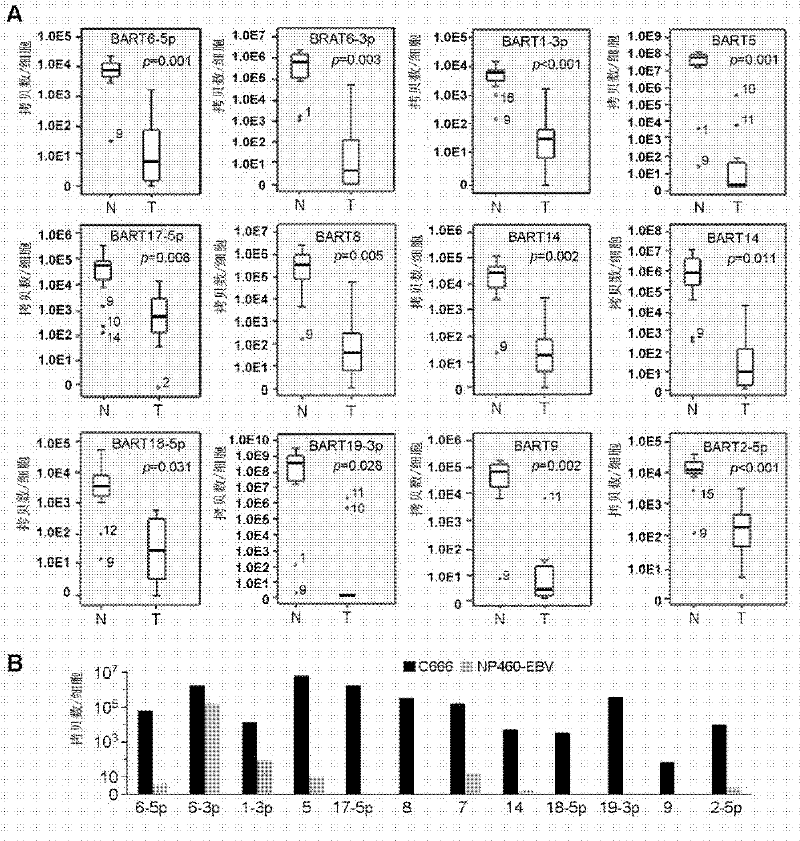
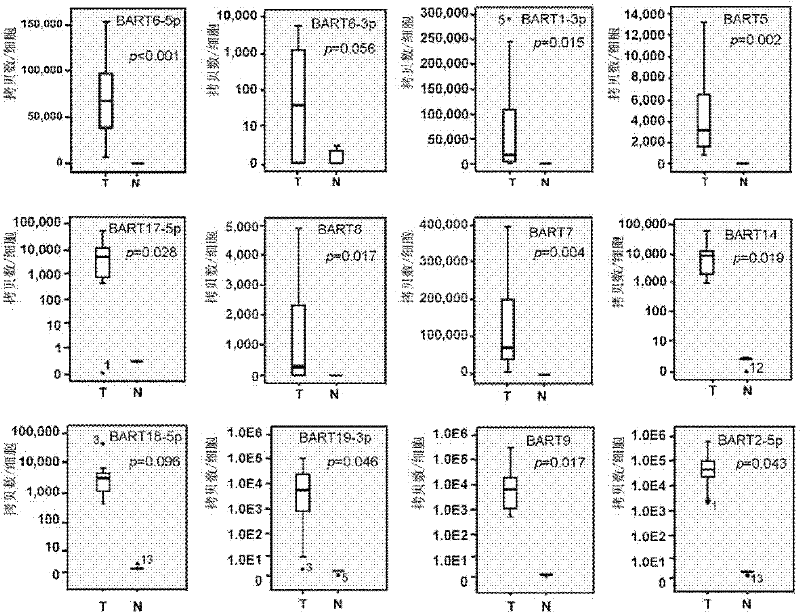
[0043] In addition to the above-mentioned miRNA chips from Exiqon, 15 pairs of tissue samples were used. These samples were 20 pairs of nasopharyngeal carcinoma tissue samples obtained from the Royal Mary Hospital in Hong Kong in 2004. The tumors were taken separately with the help of fiberoptic nasopharyngoscope Nonneoplastic tissue with normal mucosa at the site of growth and corresponding adjacent carcinoma. The total RNA in the tissue was extracted and reverse-transcribed into cDNA by real-time fluorescent quantitative PCR to detect and verify 12 Epstein-Barr virus miRNAs (including BART6-5p, 6-3p, 1-3p, 5 , 17-5p, 8, 7, 14, 18-5p, 19-3p, 9 and 2-5p) in these 15 pairs of nasopharyngeal carcinoma and corresponding normal nasopharyngeal tissues.
[0044] Total RNA was extracted by using the mirVana microRNA extraction kit (Ambion, Austin, Tex) according to its operat...
Embodiment 3
[0048] Embodiment 3: A preferred example of comparing the expression levels of the above-mentioned 12 EBV miRNAs in 15 pairs of nasopharyngeal carcinoma samples and 2 EBV-positive cell lines
[0049] 50 up-regulated miRNAs were detected by miRNA chip, and 12 EBV miRNAs with high multiples of change, high abundance and possible target genes of tumor suppressor genes were selected, among which 5 came from the EBV BART1 region (BART6-5p , 6-3p, 1-3p, 5, 17-5p), 6 from the EBV BART2 region (BART8, 7, 14, 18-5p, 9, 19-3p) and BART2-5p. The copy numbers of EBV miRNAs in each cell of these 15 pairs of NPC samples were calculated to determine whether their expression levels were sufficient to down-regulate their target genes. The calculation method of the EBV miRNA copy number in each cell is as follows: Similar to the method used in Cosmopoulos et al, C666 was diluted to a serial known concentration, and then the Ct value of EBV miRNA at each dilution concentration was detected by qP...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com