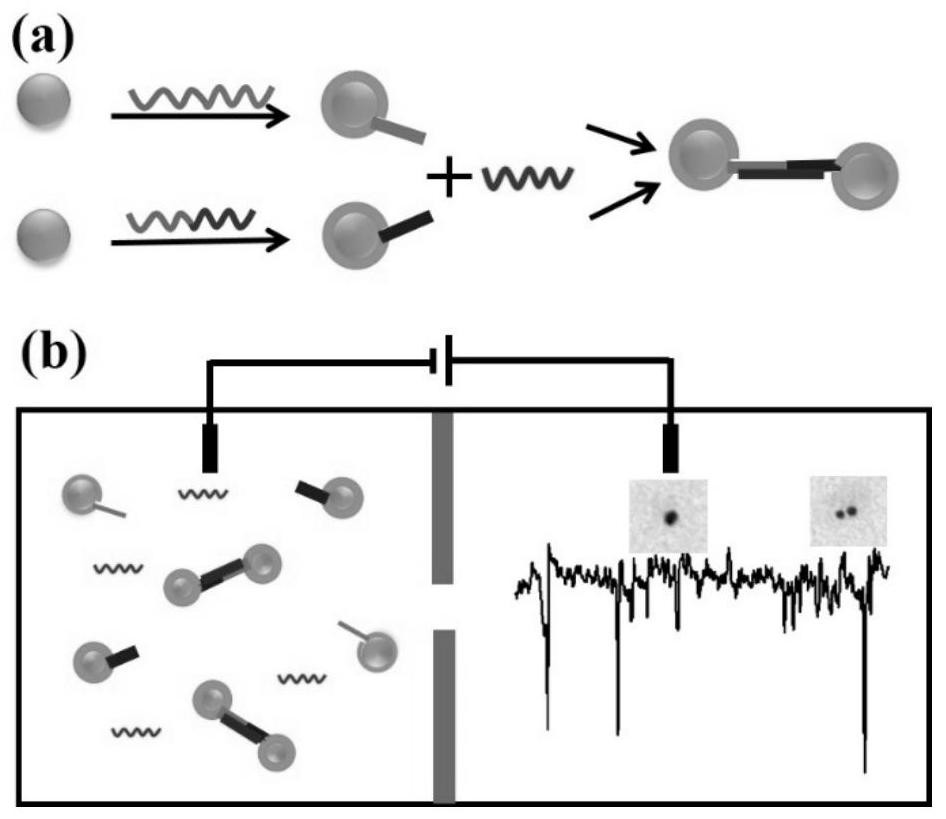
MiRNA detection method based on a solid nanopore sensor, detection probe and kit.
A technology for nanopore sensors and detection probes, which is applied in the directions of DNA/RNA fragments, recombinant DNA technology, microbial measurement/inspection, etc. It can solve the problems of difficult detection, unfavorable high-throughput detection, and high cost of nanopores Improving the effectiveness of early diagnosis and treatment
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0060] Embodiment 1: the preparation of gold nanoparticle probe and the concrete implementation to miRNA target molecular detection comprise the following steps:
[0061] Step 1: BSPP (3 mg) was added to 10 ml of AuNPs solution (the diameter of AuNPs is 5 nm), and the mixture was shaken overnight at room temperature.
[0062] Step 2: Slowly add sodium chloride to this mixture while stirring until the color changes from dark burgundy to light purple.
[0063] Step 3: The resulting mixture was centrifuged for 5 min (8000 rpm), and then the supernatant was removed. Then add 0.5ml BSPP (2.5mM / L), and add 0.5ml methanol. The mixture was centrifuged again, the supernatant was removed, and 100 uL (2.5 mM / L) of BSPP was added.
[0064] Step 4: Add the DNA fragment shown in SEQ ID NO.1 to BSPP-AuNPs for single-molecule labeling of DNA probes.
[0065] Specifically: (1) First, incubate BSPP-AuNPs with the DNA fragment shown in SEQ ID NO.1 at a molar ratio of 1:10 for 16 hours, and sh...
Embodiment 2
[0077] Example 2: Solid state nanopore detection of miR-21 and miR-486
[0078] (1) miRNA target molecules including miR-21 and miR-486 were added in equal proportions to the designed probe solution, and the mixture was left overnight.
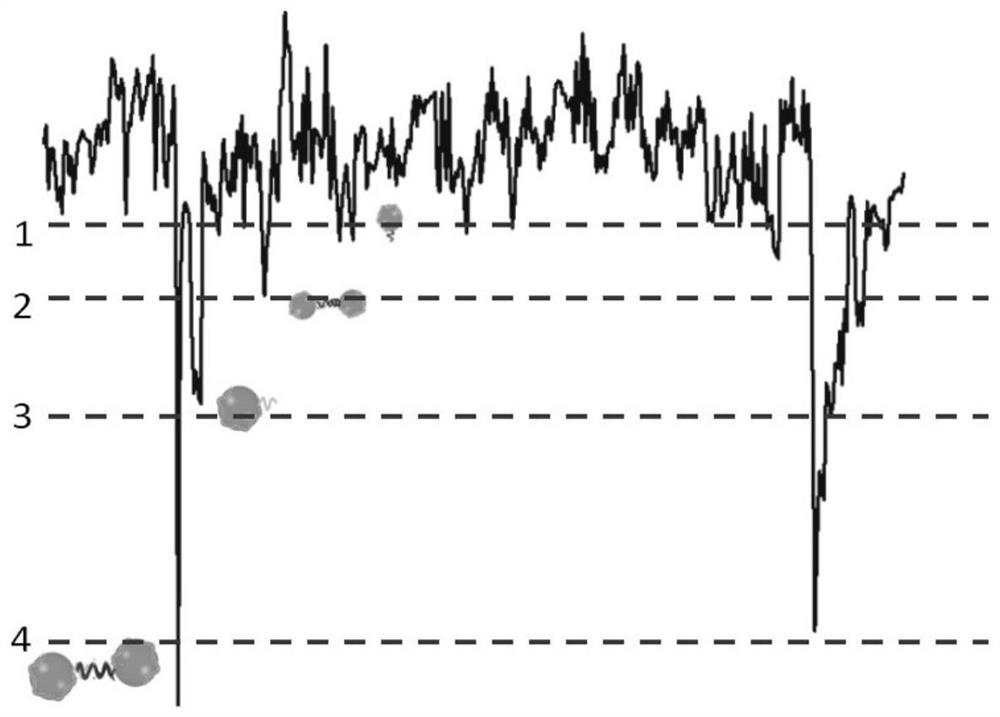
[0079] (2) Add the reacted solution to a silicon nitride solid-state nanopore sensor with a pore size of 40nm, apply different bias voltages of 0-1000mV for signal detection, drive the gold ball through the hole, observe the translocation process of the nanoparticle, and use a diaphragm Clamp records trace current signals. Its sampling frequency is 100KHz, and the cutoff frequency of low-pass filter is 10KHz.
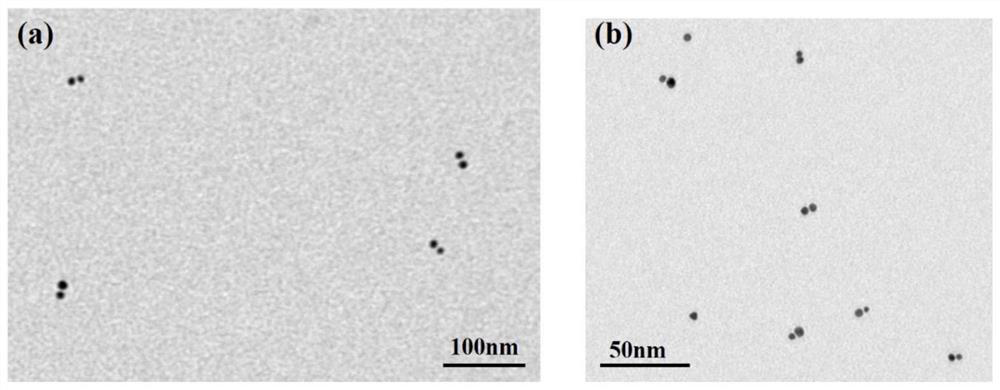
[0080] figure 2 The TEM image of the nanoparticle dimer formed after the gold nanoparticles regulated by the DNA probe bind to the miRNA target molecules, from figure 2 It can be seen from the results that the detection probe designed by the present invention has a strong adsorption capacity on the adenine A sequence at one end of ...
PUM
| Property | Measurement | Unit |
|---|---|---|
| particle diameter | aaaaa | aaaaa |
| particle diameter | aaaaa | aaaaa |
| pore size | aaaaa | aaaaa |
Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com