Target gene capable of improving regeneration capacity of plants, regulation molecules and application of target gene
A regenerative capacity, plant technology, applied in the field of botany, can solve the problem of not meeting the needs of explaining the mechanism of plant regeneration
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment approach
[0072] As an embodiment of the present invention, the expression of the HAM gene in plants can be down-regulated by knocking out the HAM gene.
[0073] As an embodiment of the present invention, the CRISPR / Cas9 system can be used for gene editing, thereby knocking out the HAM gene and improving plant regeneration ability. Appropriate sgRNA target sites will lead to higher gene editing efficiency, so it is more important to design and find appropriate target sites before proceeding with gene editing. After designing specific target sites, in vitro cell activity screening is required to obtain effective target sites for subsequent experiments.
[0074] As an embodiment of the present invention, virus-induced gene silencing (VIGS) can be used to inhibit HAM, thereby improving plant regeneration ability.
[0075] It should be understood that after knowing the correlation between HAM and plant traits, those skilled in the art can prepare molecules that down-regulate HAM in various...
Embodiment 1
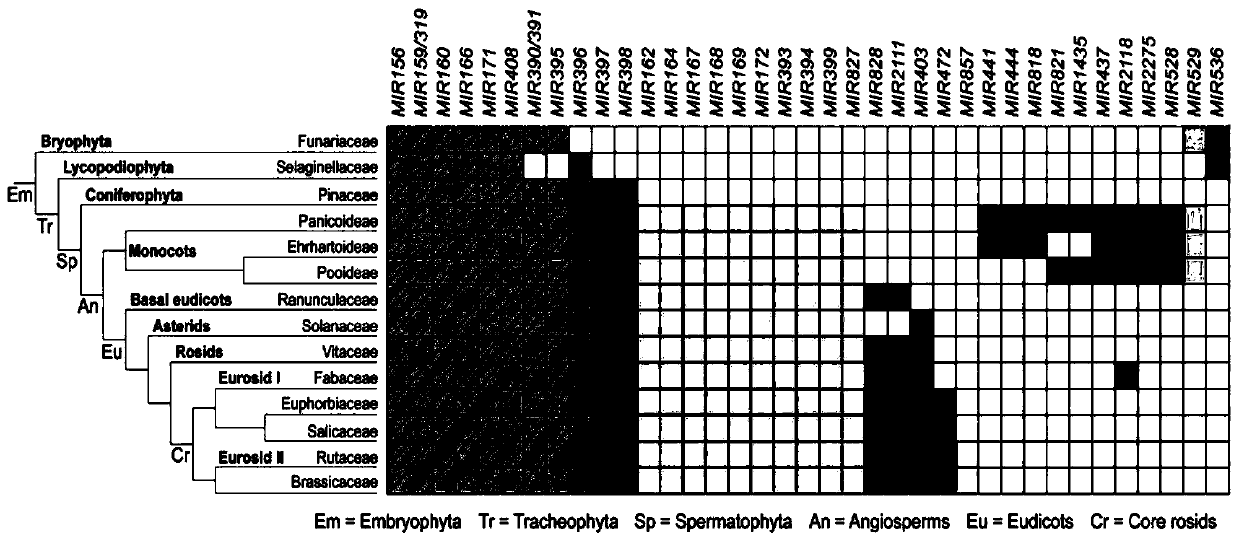
[0143] Example 1, miR171 and its target gene HAM are conserved in all plants
[0144] The target genes of miR171 are GRAS transcription factors, including three genes in the Arabidopsis genome, namely HAM1, HAM2 and HAM3.
[0145] Evolutionary analysis showed that miR171 is conserved in all terrestrial plants and widely exists in bryophytes, ferns, gymnosperms and angiosperms, such as figure 1 .
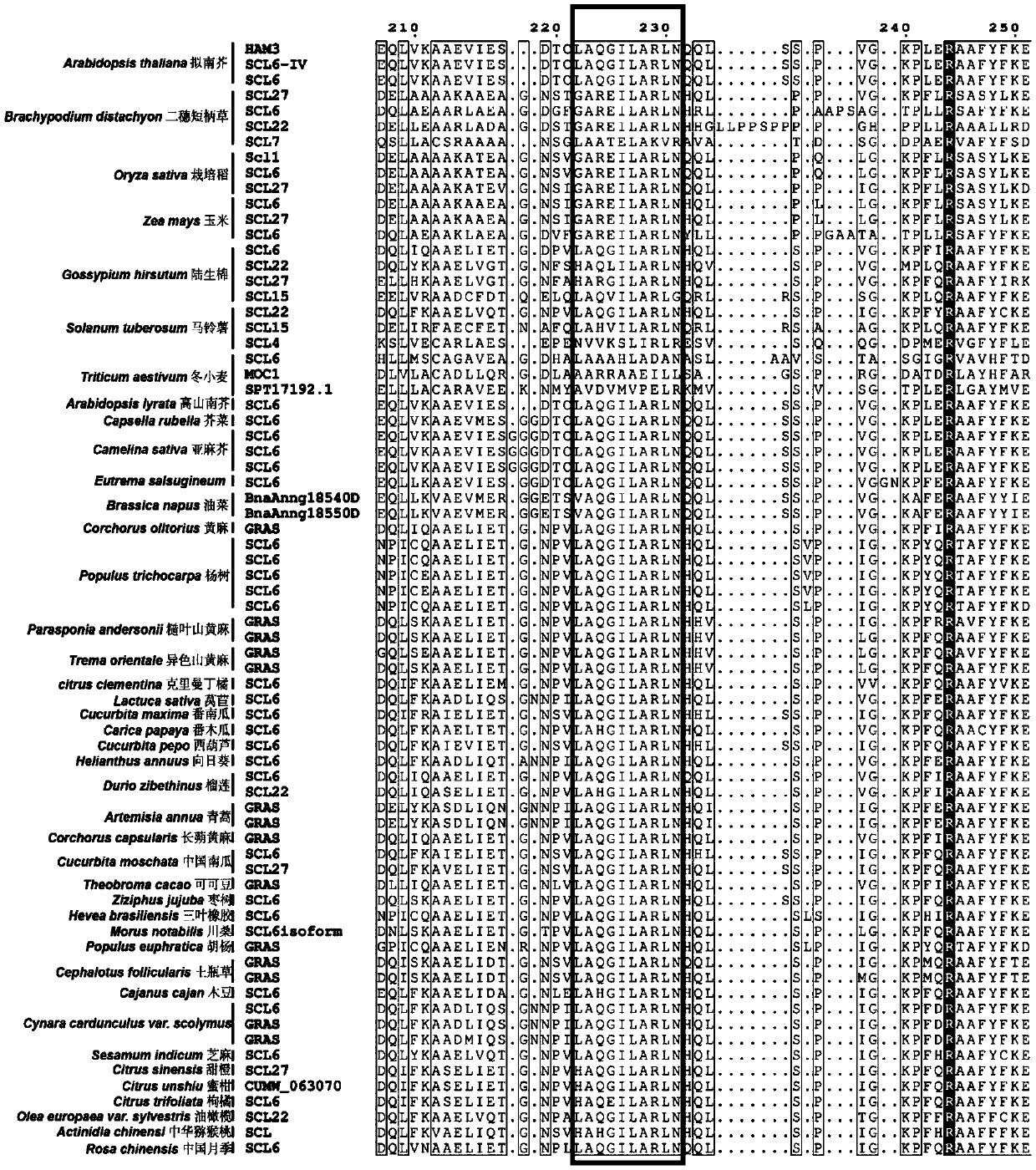
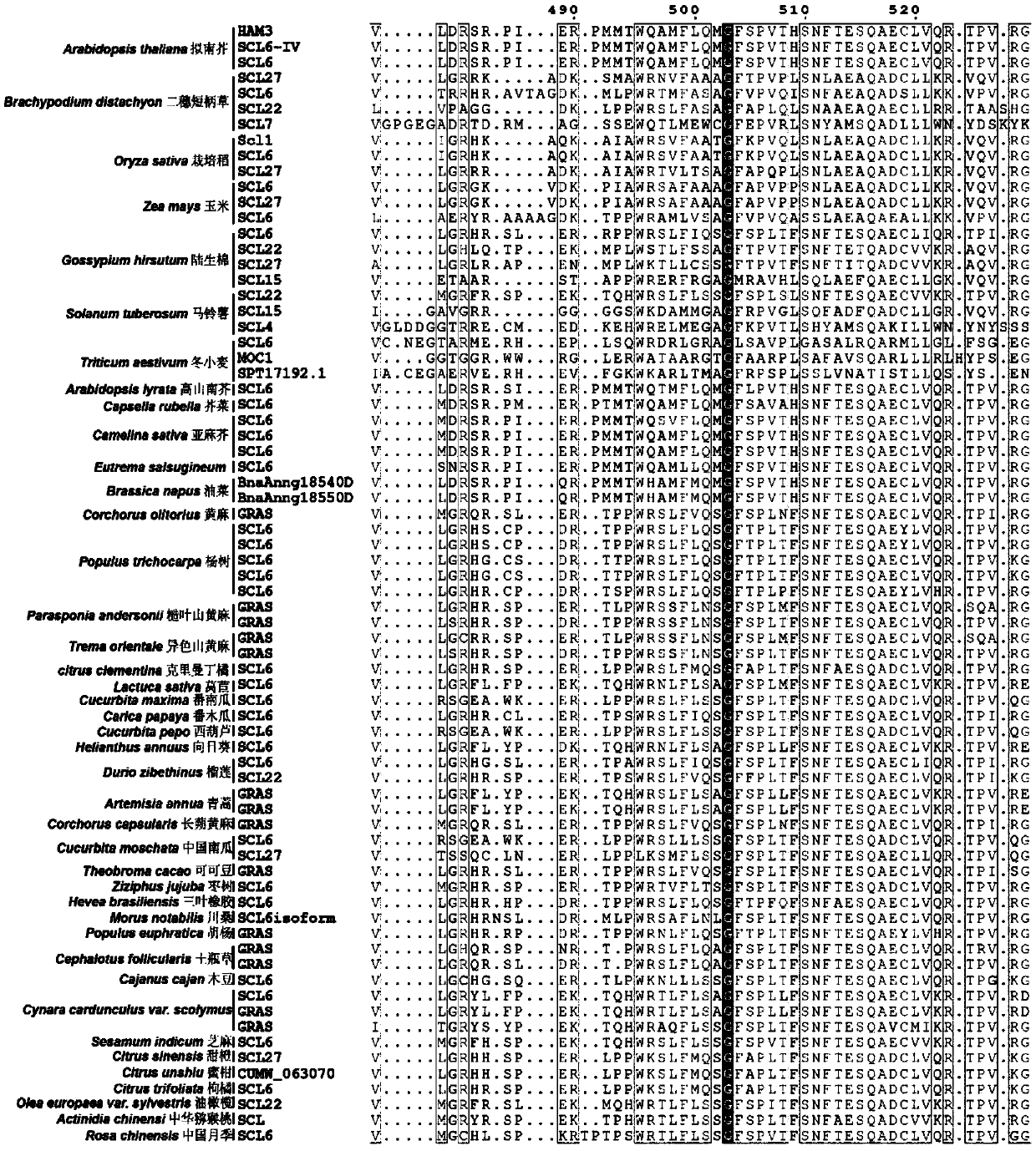
[0146] At the same time, the present inventors used the protein sequence "blast" function of the NCBI website (https: / / blast.ncbi.nlm.nih.gov / Blast.cgi?PROGRAM=blastp&PAGE_TYPE=BlastSearch&LINK_LOC=blasthome) to search for the complete sequence of HAM3 in Arabidopsis Long protein sequence, obtained protein sequences highly homologous to the Arabidopsis HAM3 protein sequence in several different species, and obtained the sequence information through MAFFT software (https: / / mafft.cbrc.jp / alignment / software / ) analysis After obtaining the homology information, some homology information...
Embodiment 2
[0147] Example 2, miR171 regulates Arabidopsis shoot regeneration ability
[0148] There are three coding genes for miR171 in the Arabidopsis genome, MIR171A, MIR171B, and MIR171C; their coding products, namely mature sequences, are miR171a, miR171b, and miR171c, respectively, and the mature sequences of miR171b and miR171c are the same. The present inventors prepared mir171a, mir171b and mir171ab mutants respectively, and investigated the shoot regeneration ability of the mutants.
[0149] The result is as image 3 As shown, the shoot regeneration ability of mir171b and mir171a b was significantly reduced, suggesting that miR171 is mainly involved in the regulation of shoot regeneration ability.
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com