Recombinant DNA constructs and methods for modulating expression of a target gene
A technology of constructs and target genes, applied in the direction of recombinant DNA technology, biochemical equipment and methods, DNA/RNA fragments, etc., can solve the problems of no reports
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
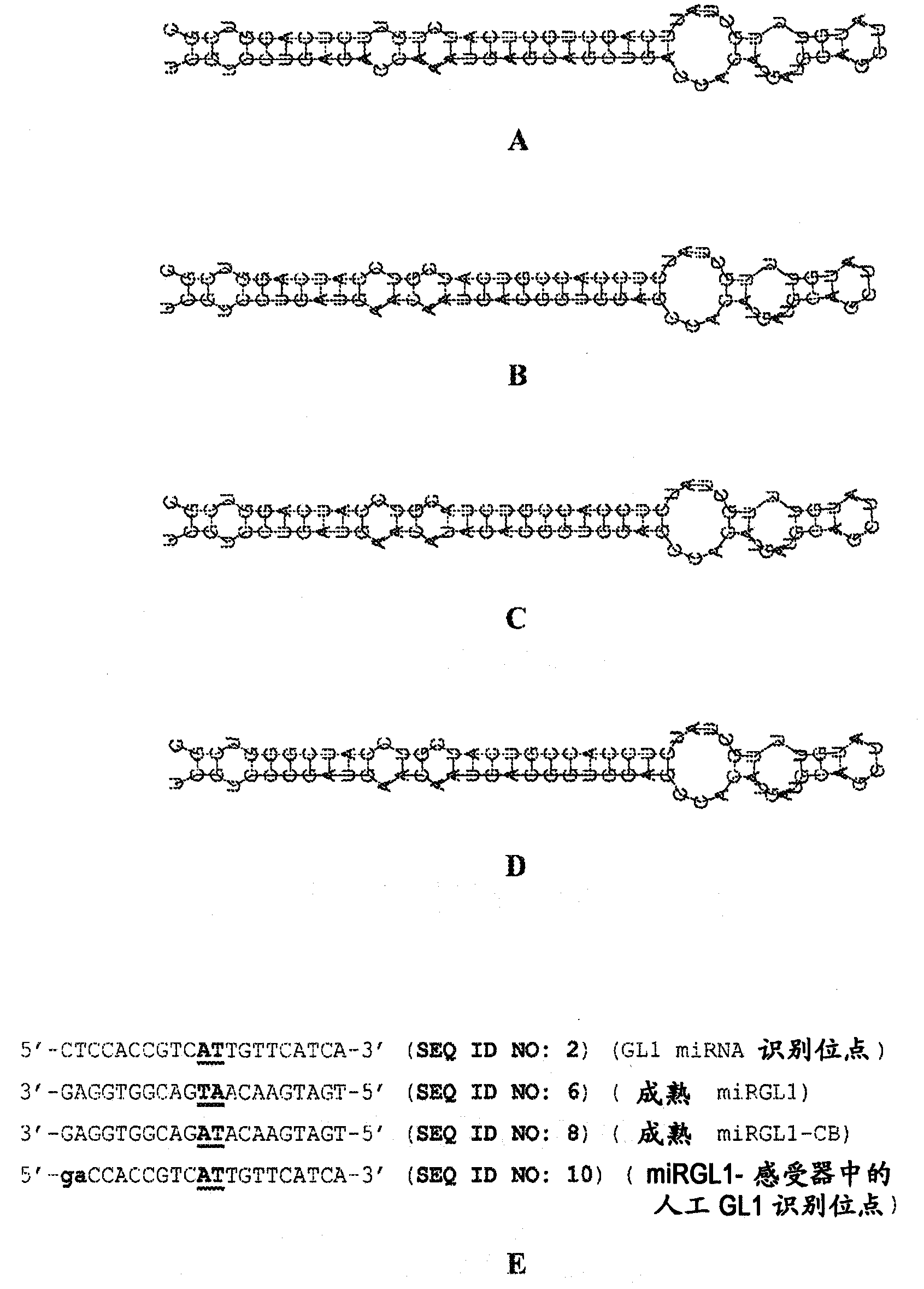
[0162] This example illustrates the preparation and use of "cleavage blocker" recombinant DNA constructs containing DNA processed into RNA containing single-stranded RNA, the single-stranded RNA and the transcript of at least one target gene A hybrid fragment of RNA that binds to form at least partially double-stranded RNA that renders the transcript resistant to cleavage by RNase III ribonucleases in or near the hybrid fragment; wherein the binding of the single-stranded RNA to the transcript ( and eventual formation of hybrid fragments) inhibits double-stranded RNA-mediated repression of target genes. More specifically, this example describes constructs for the production of artificial or genetically engineered miRNAs or cleavage blockers in plants, and which cleavage blockers inhibit miRNA-mediated miRNA-mediated transformation of the Arabidopsis GL1 gene in transformed plant cells. The use of induced inhibition.
[0163] Target gene: The Arabidopsis GLABROUS1 (GL1) gene...
Embodiment 2
[0176] This example illustrates an alternative "cleavage blocker" recombinant DNA construct with a modification at the position corresponding to the 5' end of the mature miRNA that naturally binds to the recognition site of the target gene, i.e., a "cleavage blocker" produced transgenic in plants. 5'-modified cleavage blockers", and methods of using the cleavage blockers to inhibit miRNA-mediated suppression of target genes in transformed plant cells.
[0177] In one example, DNA encoding an artificial miRNA (miRGL1) precursor (SEQ ID NO: 6) was modified by a single nucleotide change (changing the 5' end of mature miRGL1 from U to C), resulting in a 5'- Modified cleavage blocker precursor sequence AATTCATTACATTGATAAAACACAATTCAAAAGATCAATGTTCCACTTCATGCAAAGACATTTCCAAAATATGTGTAGGTAGAGGGGTTTTACAGGATCGTCC CGATGAACAATGACGGTGGAG CCACATGATGCAGCTATGTTTGCTAT GGTCGCCCTTGTTGGACTGTCCAACTCCTACTGATTGCGGATGCACTTGCCACAAATGAAAATCAAAGCGAGGGGAAAAGAATGTAGAGTGTGACTACGATTGCATGCATGTGATTTAGGTAATTAAG...
Embodiment 3
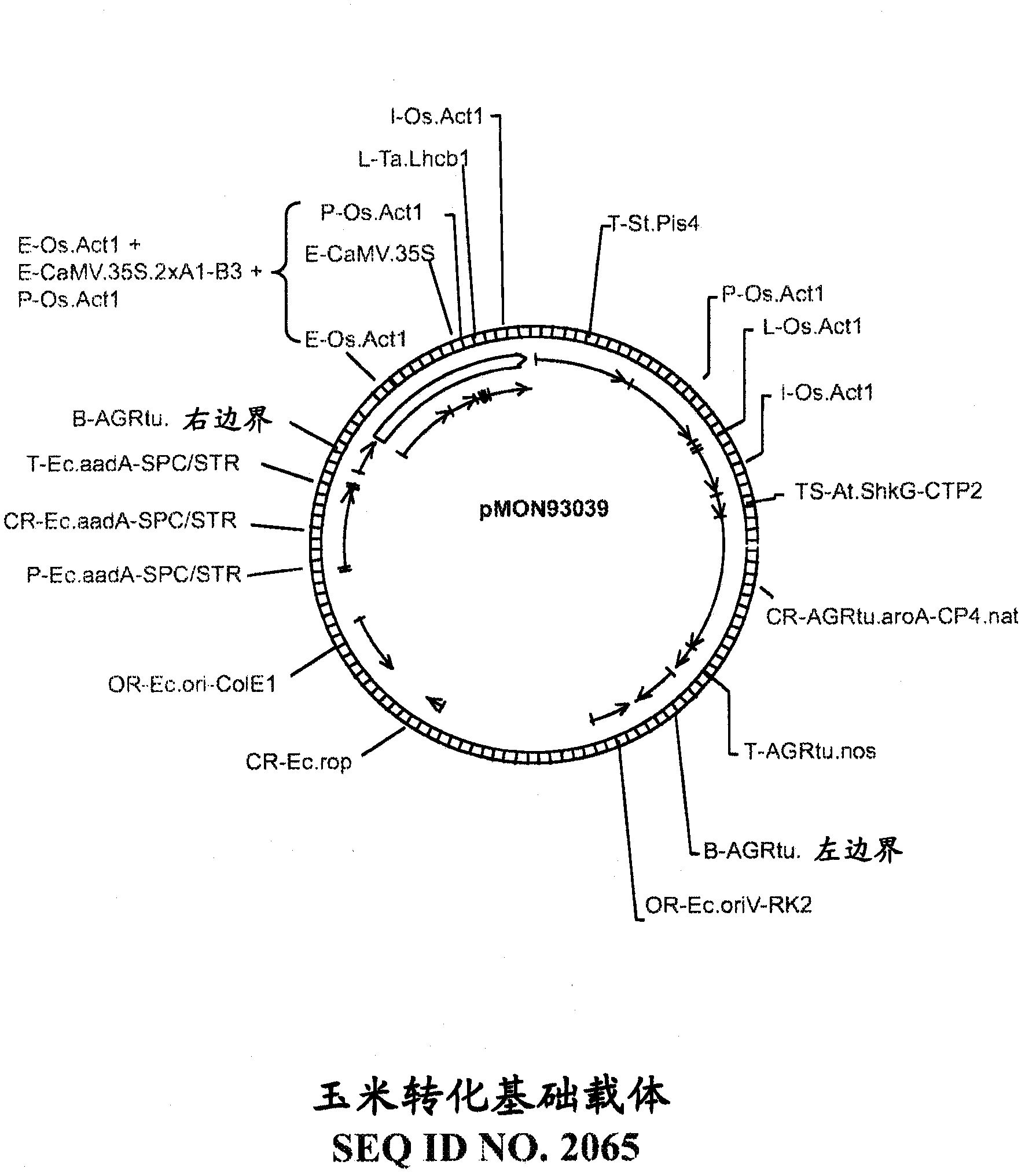
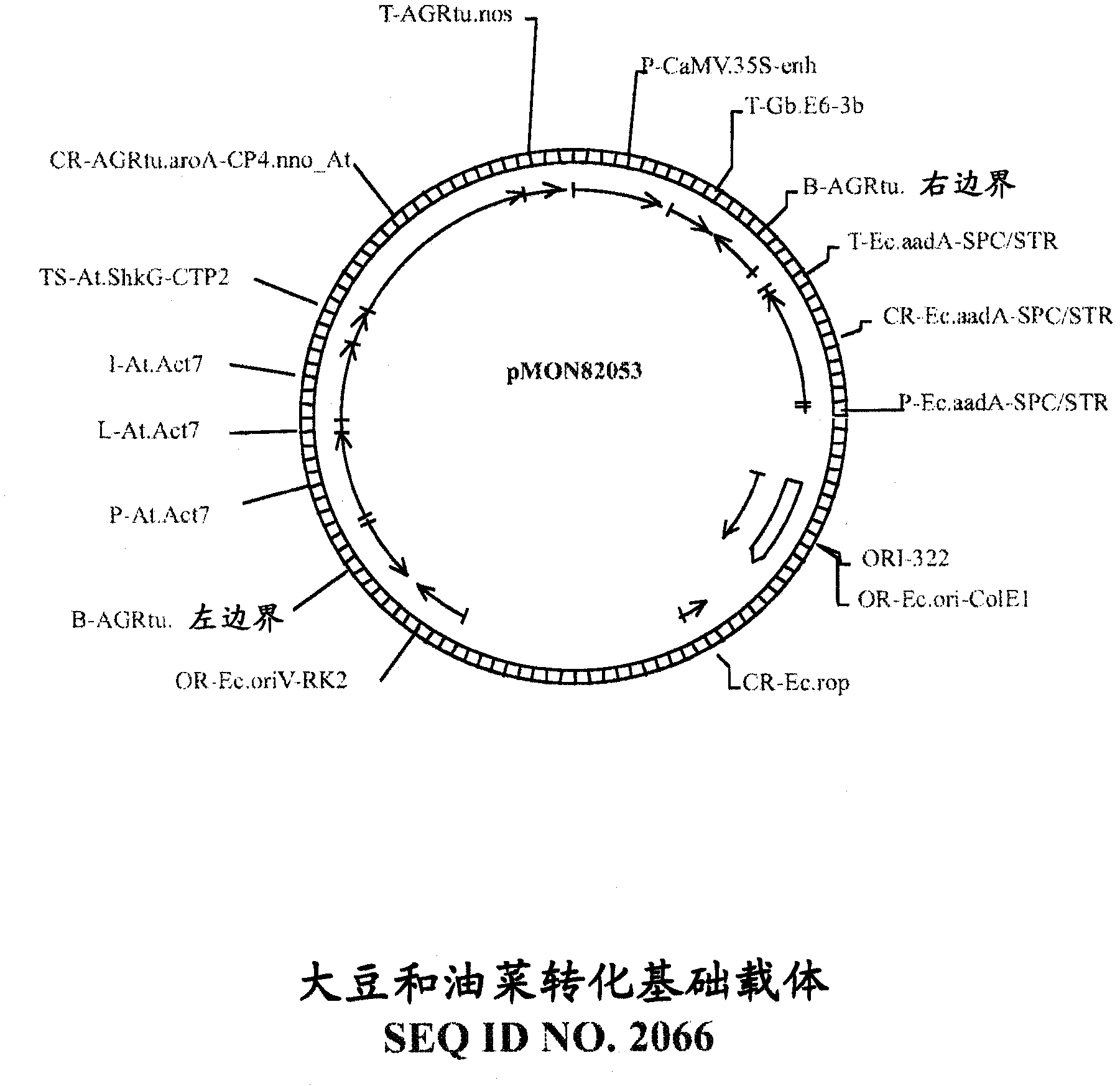
[0184] This example provides an embodiment of a target gene identified as a "verified miRNA target" (ie, containing a verified miRNA recognition site). The recombinant DNA constructs of the present invention can be used to modulate the expression of such target genes and can be used to prepare non-naturally transgenic plant cells, plant tissues and plants (particularly non-naturally transgenic crops) with increased yield or other desirable traits.
[0185] Prediction of recognition sites is accomplished using methods known in the art, eg, the sequence complementarity rules described by Zhang (2005) Nucleic Acids Res., 33:W701-704 and Rhoades et al. (2002) Cell, 110:513-520. One method of experimentally confirming predicted miRNA recognition sites is a technique known as RNA ligase-mediated rapid amplification of the 5' end of cDNA ("5'RLM-RACE" or "5'RACE"), which identifies miRNA cleavage patterns; see eg Kasschau et al. (2003) Dev. Cell, 4:205-217 and Llave et al. (2002) Sci...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com