Index acceleration method and corresponding system in scale protein identification
A protein identification and protein technology, applied in special data processing applications, instruments, electrical digital data processing, etc., can solve problems such as unreasonable division of temporary files, uneven distribution of peptide sequences, defects in time and space performance, etc., to achieve effective Conducive to storage and query, low space consumption, and improved efficiency
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment Construction
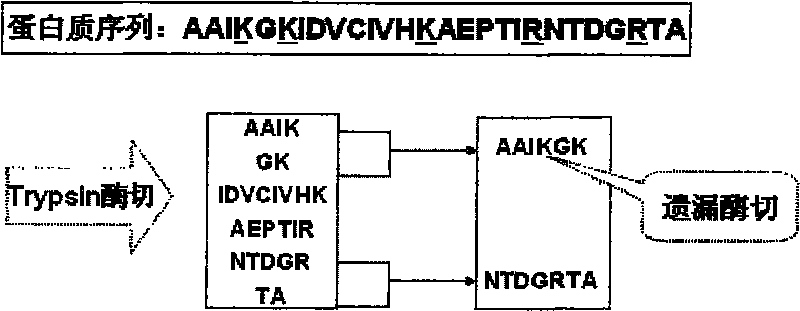
[0061] Before the present invention is described, explain some nouns involved in the present invention:
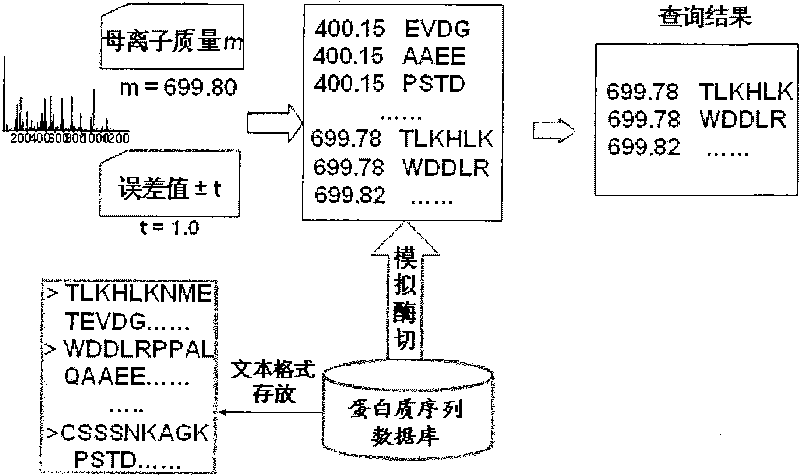
[0062] Peptide Sequence Dictionary: A file for storing non-repetitive peptide sequences. Generally speaking, there are usually two ways to store peptide sequences: first, store them as actual sequences; second, store them as structures with fixed-length characteristics, including pointers and lengths, and pointers point to peptide sequences that appear in the original database The position of , the length indicates the length of the peptide sequence through which the peptide sequence can be read in the protein database. The peptide sequence dictionary of the present invention stores peptide sequence information in the second storage mode and includes a pointer to a posting list.
[0063] Posting table: A file used to store correspondences between peptide sequences and proteins. A peptide sequence may appear in several proteins, and a record in the posting table stores th...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com