Method for preparing hydroxyalkanoate homopolymer and special bacteria thereof
A technology of polyhydroxyalkanoate and coding gene, which is applied in the field of preparing hydroxyalkanoate homopolymer, and can solve the problems of undiscovered wild strains of microorganisms and the like
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0087] Embodiment 1, construction and biosynthesis of mutant strain Pseudomonas putida KTOY08
[0088] 1. Construction of mutant strain Pseudomonas putida KTOY08
[0089] (1) Construction of suicide plasmid pSPK09
[0090] 1) Using the genomic DNA of Pseudomonas putida strain KT2442 as a template, under the guidance of primers P1: ATTCTAGAGCTGAACCCGGATGGC and P2: AATAAGCTTGCCGAAGCGCAGGATC, PCR amplified two connected genes, 3-ketoacyl-CoA thiolase gene (fadAx) and 3-ketoacyl-CoA thiolase gene (fadAx) -Hydroxyacyl-CoA dehydrogenase gene (fadB2x), the PCR amplification product was double-digested with restriction endonucleases XbaI and HindIII, and the digested fragment with a size of about 2.7kb was recovered.
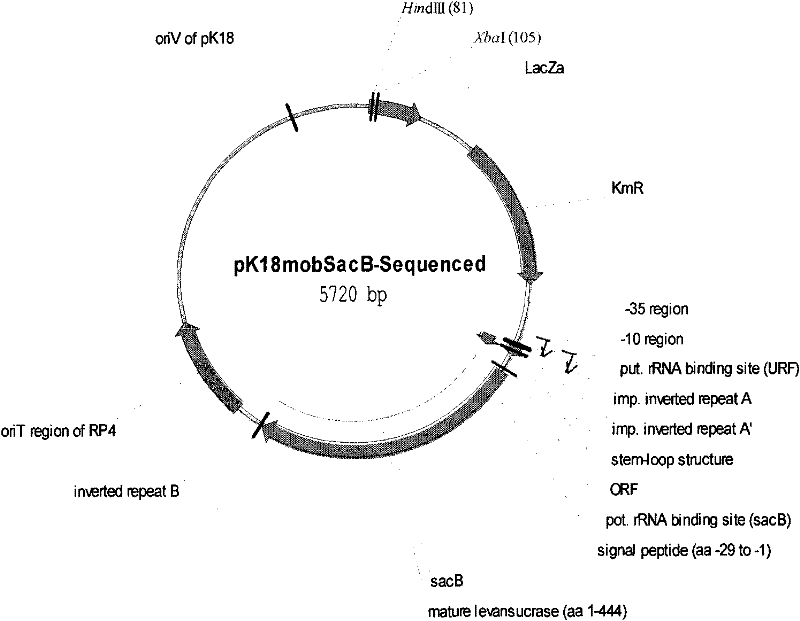
[0091] 2) Carry out double digestion of plasmid pK18mobSacB with restriction endonucleases XbaI and HindIII, and recover a digested fragment with a size of about 5.699 kb;
[0092] 3) Ligate the recovered fragments of step 1) and step 2) with T4DNA ligase, and after t...
Embodiment 2
[0137] Example 2, construction and biosynthesis of bacterial strain Pseudomonas putida KTOY08-G
[0138] 1. Construction of engineering strain Pseudomonas putida KTOY08-G
[0139] (1) Construction of recombinant vector pKQQ01
[0140] 1) Using the genomic DNA of Pseudomonas putida KT2442 as a template, PCR amplification was carried out with primers phaG-up-1 and phaG-up-2 to obtain the 3-hydroxyacyl apolipoprotein-CoA transferase gene (3-hydroxyacyl- The upstream fragment of acylcarrier protein-coenzyme A transferase (or PhaG) (sequence 3 in the sequence listing is shown in the 1st-448th nucleotide from the 5' end); use restriction endonucleases XbaI and HindIII for PCR amplification products Carry out double enzyme digestion, and recover the enzyme-cut fragment with a size of about 400bp;
[0141]Using the genomic DNA of Pseudomonas putida KT2442 as a template, PCR amplification was carried out with primers phaG-down-1 and phaG-down-2 to obtain the gene encoding 3-hydroxyac...
Embodiment 3
[0189] Example 3, construction of strain Pseudomonas putida KTHH06 and biosynthesis of PHA
[0190] Vector pKSSE5.3 (Hein S, B, Gottschalk G, Steinbüchel A. Biosynthesis of poly(4-hydroxybutyric acid) by recombinant strains of Escherichia coli. FEMS Microbiol. Lett. 1997; 153: 411-418.) (Provided by Tsinghua University);
[0191] The vector pKSSE5.3 contains the poly-β-hydroxybutyrate synthase coding gene (phbC gene) from the Ralstonia eutropha genome; the poly-β- The nucleotide sequence of the gene encoding hydroxybutyrate synthase (phbC gene) is shown as sequence 4 in the sequence listing, which is 1767bp.
[0192] Vector pBBR1-MCS2 (Michael E. Kovach, Philip H. Elzer, D. Steven Hill et al. Four new derivatives of the broad-host-range cloning vector pBBR1MCS, carrying different antibiotic-resistance cassettes. Gene. 1995; 166: 175-176 .) (provided by Tsinghua University);
[0193] The vector pBBR1-MCS2 contains the 4-hydroxybutyryl-CoA transferase encoding gene (orfZ gen...
PUM
| Property | Measurement | Unit |
|---|---|---|
| concentration | aaaaa | aaaaa |
| concentration | aaaaa | aaaaa |
| concentration | aaaaa | aaaaa |
Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com