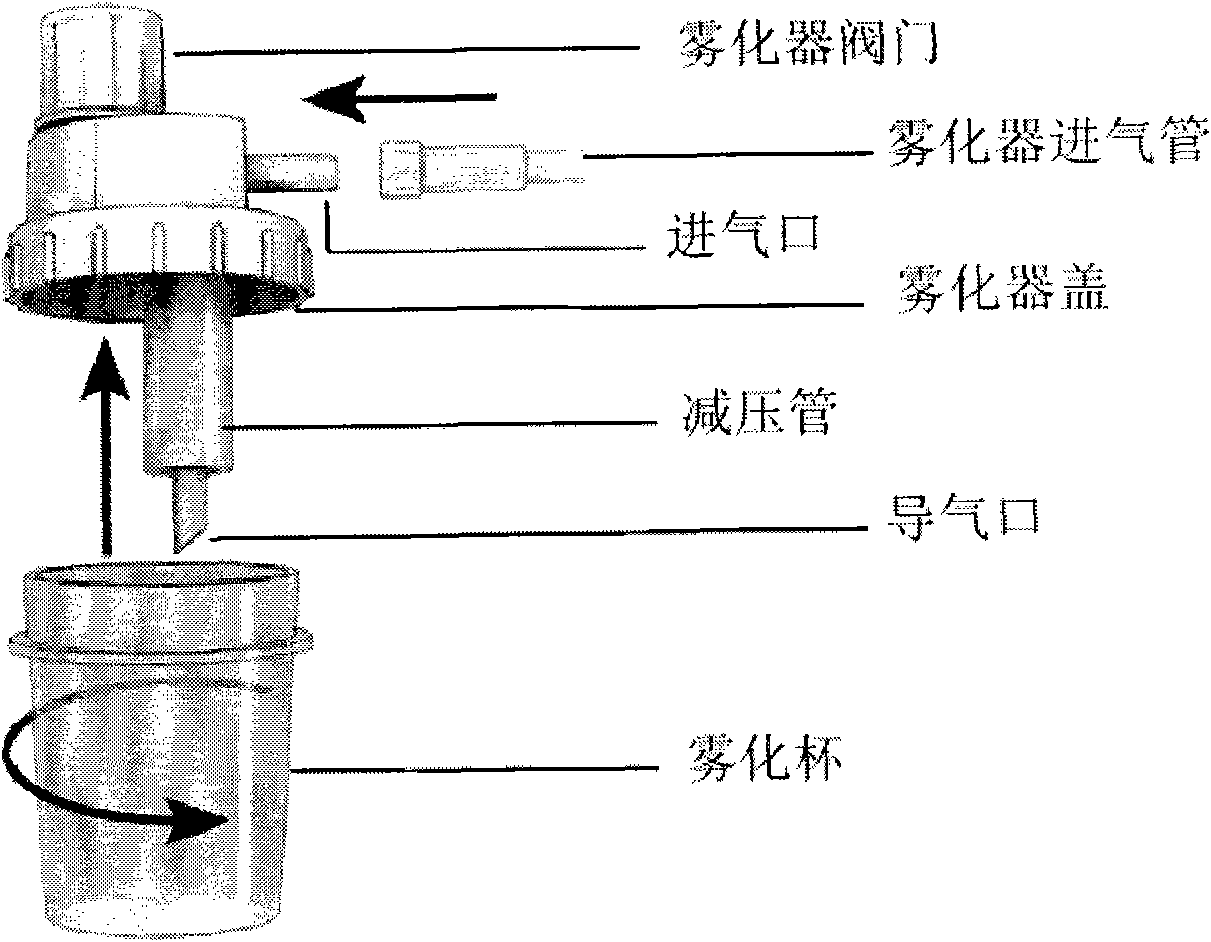
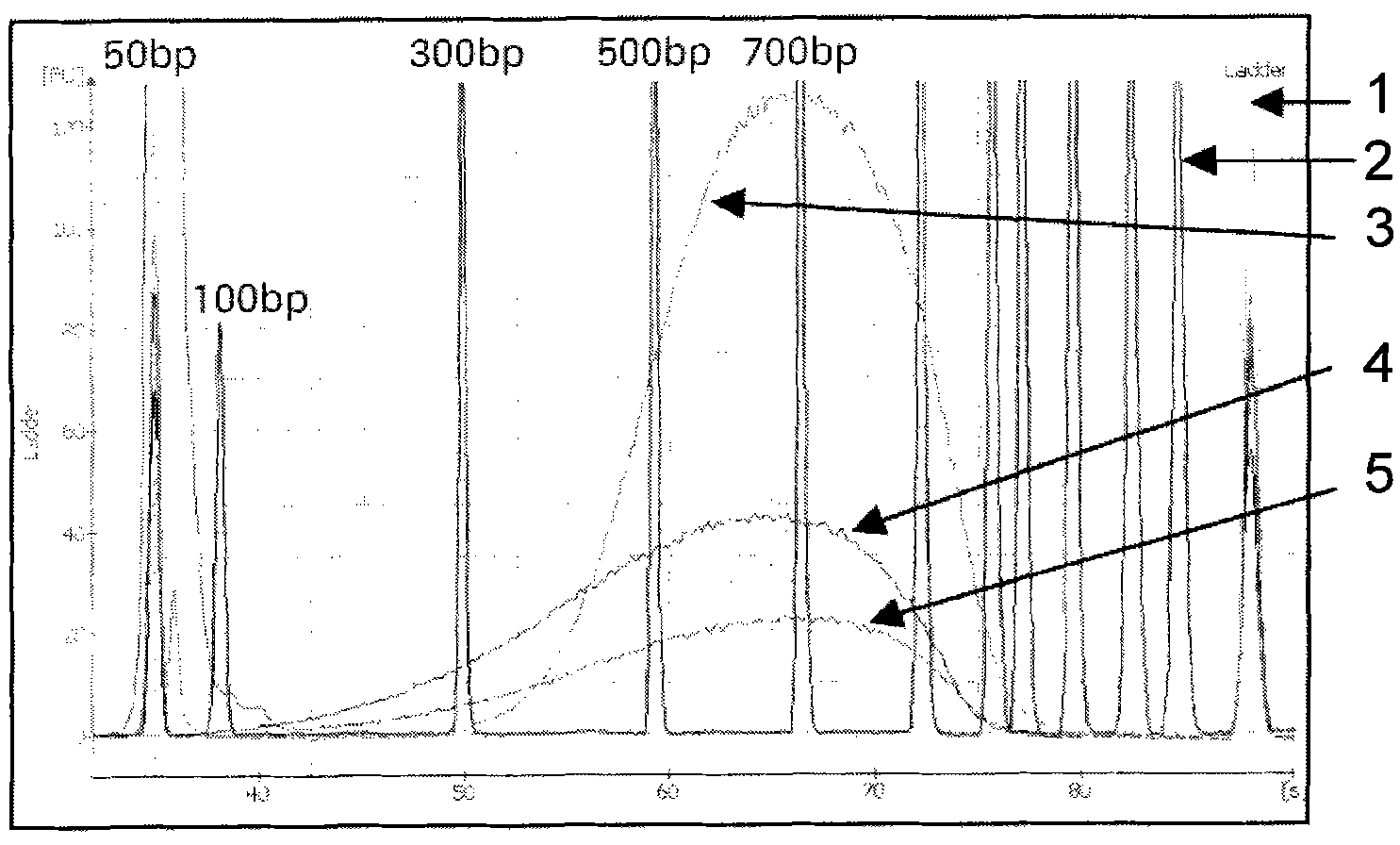
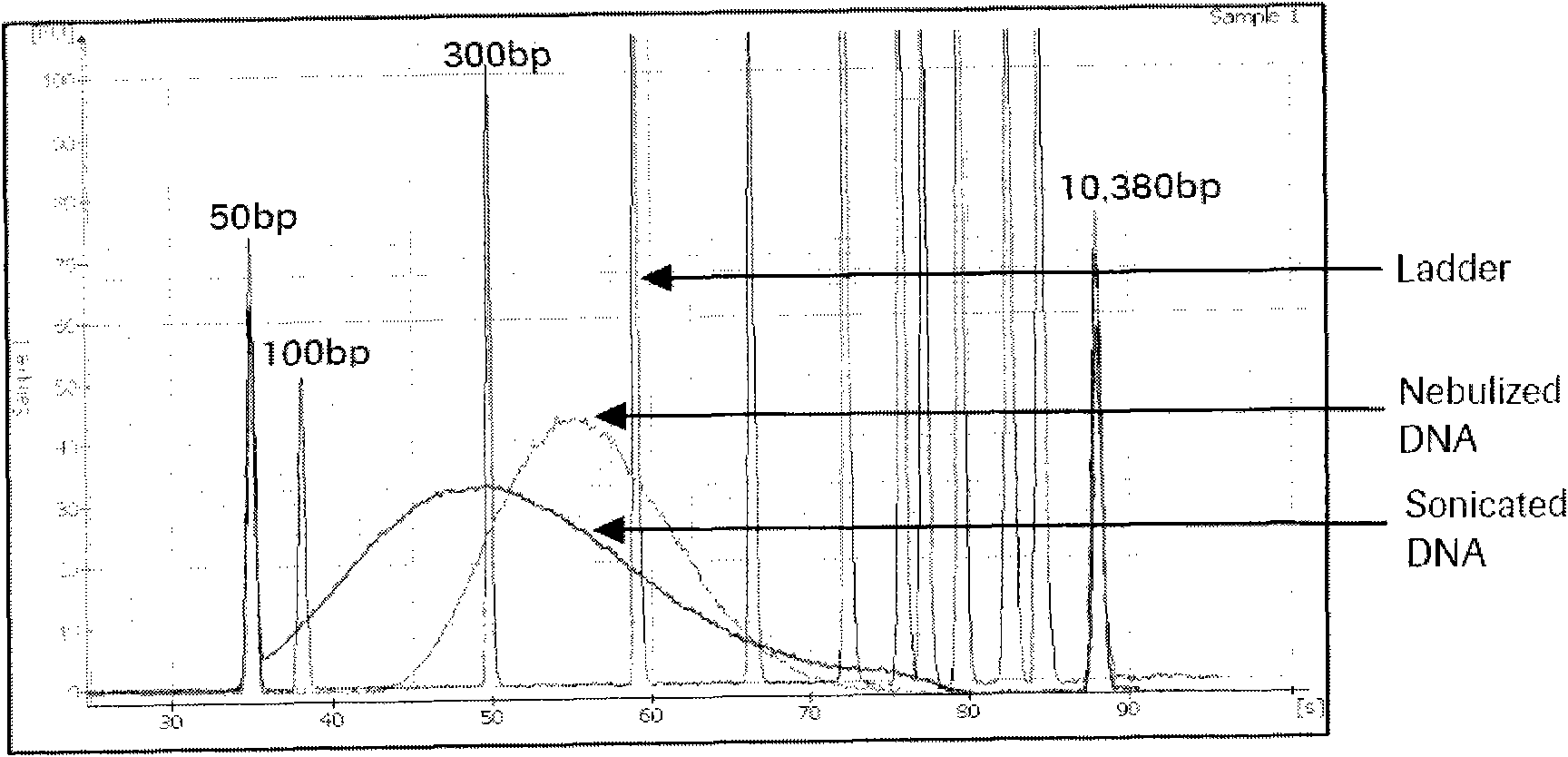
Method for correcting sequence abundance deviation of secondary high-flux sequence test by DNA sequence bar codes
A DNA-sequencing, high-throughput technology, applied in biochemical equipment and methods, microbial assay/inspection, etc., to solve problems such as contamination, water-in-oil globule rupture, false abundance, etc.
Inactive Publication Date: 2010-09-29
SUZHOU ZHONGXIN BIOTECH
View PDF2 Cites 12 Cited by
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
A major defect of the emulsion PCR technology is that the water-in-oil pellets inevitably rupture during the PCR reaction and contaminate other blank pellets, which leads to falsely increas
Method used
the structure of the environmentally friendly knitted fabric provided by the present invention; figure 2 Flow chart of the yarn wrapping machine for environmentally friendly knitted fabrics and storage devices; image 3 Is the parameter map of the yarn covering machine
View moreImage
Smart Image Click on the blue labels to locate them in the text.
Smart ImageViewing Examples
Examples
Experimental program
Comparison scheme
Effect test
 Login to View More
Login to View More PUM
 Login to View More
Login to View More Abstract
The invention relates to a method for correcting the sequence abundance deviation of secondary high-flux sequence test by DNA sequence bar codes. Through introducing ten or more than ten DNA sequences to be used as bar code labels, the DNA of various different samples is corrected through abundance data obtained through the high-flux sequence test. The invention introduces the DNA sequence bar codes through three different designs, and the three methods can be applicable to various different sample conditions. During the three technical designs are described, the design uses 454 high-flux sequence test as the example. However, the principle is also applicable to other high-flux sequence test technical platforms, including SOLiD, Polynator and the like. The invention effectively overcomes the experimental error introduced by the emulsion PCR steps in building databases of several platforms of the current secondary high-flux sequence test. The technical method and the experimental flow process designed by the invention distinguish and correct the higher sequence read-out abundance of some template sequence in the emulsion PCR process caused by crack pollution of water-in-oil small balls through introducing more than one million kinds of (410=1048576) DNA sequence bar codes.
Description
Technical field: [0001] The invention relates to a method for correcting the sequence abundance deviation of the second-generation high-throughput sequencing by using a DNA sequence barcode. Background technique: [0002] High-throughput sequencing technology is a revolutionary change to traditional sequencing. It can sequence hundreds of thousands to millions of DNA molecules at a time, making it possible to analyze the transcriptome and genome of a species in detail. [0003] The development of next-generation high-throughput sequencing technology started with Genome Sequencer20 System (1), an innovative ultra-high-throughput genome sequencing system based on pyrosequencing and emulsion PCR, launched by 454 at the end of 2005. In 2007, 454 launched a second-generation genome sequencing system with better performance: Genome Sequencer FLX System. At present, besides the 454 GS system, the second-generation high-throughput sequencing technology platforms that have been comm...
Claims
the structure of the environmentally friendly knitted fabric provided by the present invention; figure 2 Flow chart of the yarn wrapping machine for environmentally friendly knitted fabrics and storage devices; image 3 Is the parameter map of the yarn covering machine
Login to View More Application Information
Patent Timeline
 Login to View More
Login to View More IPC IPC(8): C12Q1/68C12Q1/70
Inventor 李轩熊慧郝沛李亦学
Owner SUZHOU ZHONGXIN BIOTECH
Features
- R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
Why Patsnap Eureka
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Social media
Patsnap Eureka Blog
Learn More Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com