Fully human anti-VEGF antibodies and methods of using
A technology of antibodies and antigens, which is applied in the field of treatment of medical diseases such as cancer, and can solve problems that are difficult to achieve
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0460] Example 1: Binding hVEGF 165 The production of antibodies:
[0461] For fixed hVEGF 165 Panning of human single-chain Fv (scFv) phage display library to identify binding hVEGF 165 A set of antibody fragments of ability. Use standard protocols for panning (see, for example, Methods in Molecular Biology, vol.178: Antibody Phage Display: Methods and Protocols Edited by PMO'Brien and R. Aitken, Humana Press; "Panning of AntibodyPhage-Display Libraries," Coomber. DWJ, PP. 133-145, and "Selection of Antibodies Against Biotinylated Antigens," Chames, P. et al., pp. 147-157).
[0462] Briefly, use 50μl of recombinant hVEGF at a concentration of 10μg / ml in PBS 165 (R&D Systems, catalog number: 293-VE) Coating Three holes of the MAXISORP board. After overnight incubation at 4°C, free binding sites were blocked with 5% milk PBS solution for 1 hour at room temperature. Then, about 200 μl of the phage library in 5% milk / PBS was added to the closed wells and incubated at room tempe...
Embodiment 2
[0464] Example 2: Targeting hVEGF by scFv 165 Blocking of VEGF receptor binding:
[0465] Use microplate-based competitive screening Assay (Perkins Elmer, Waltham, MA), the ELISA in Example 1 showed that it is 165 Bound phage clones test them to block hVEGF 165 The ability to bind to VEGF-R1 and / or VEGF-R2.
[0466] Briefly, the biotinylated hVEGF 165 The solution was added to the cytoplasmic extract of Example 1 in a volume ratio of 1:1 to reach a final concentration of 0.5 μg / ml. Add 100 μl of this mixture to VEGF-R1 or VEGF-R2 (R&D Systems: VEGF-R1 / Flt-1, catalog number: 321-FL; VEGF-R2 / KDR / Flk-1, catalog number: 357-KD) Coated plates and incubate at room temperature for 1.5 hours. The plate was washed with PBST, and a 1:250 dilution of europium-streptavidin in DELFIA assay buffer was added in an amount of 50 μl / well. The plate was incubated for 1 hour at room temperature and then washed with DELFIA washing buffer. DELFIA enhancement buffer was added in an amount of 50 μ...
Embodiment 3
[0467] Example 3: Conversion of scFv to scFv-Fc and IgG
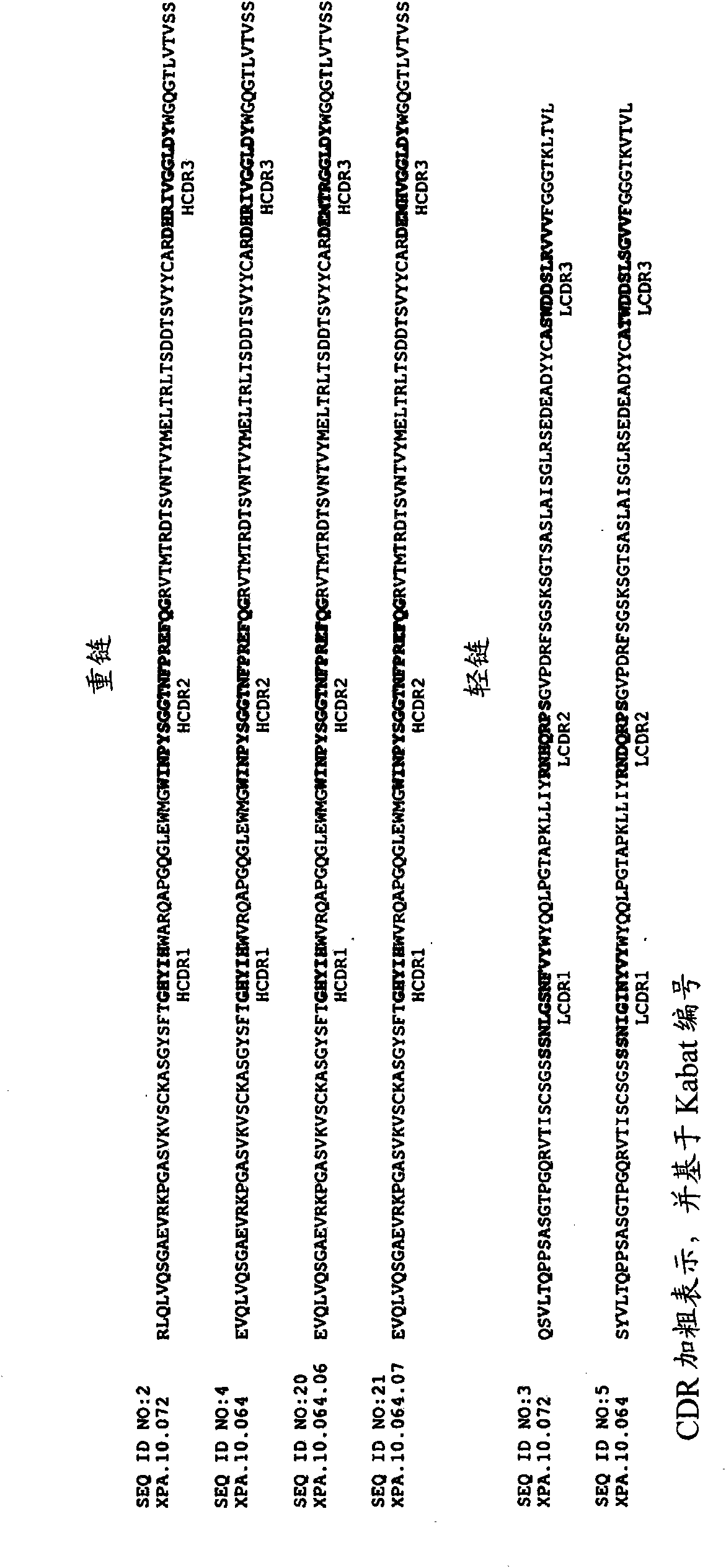
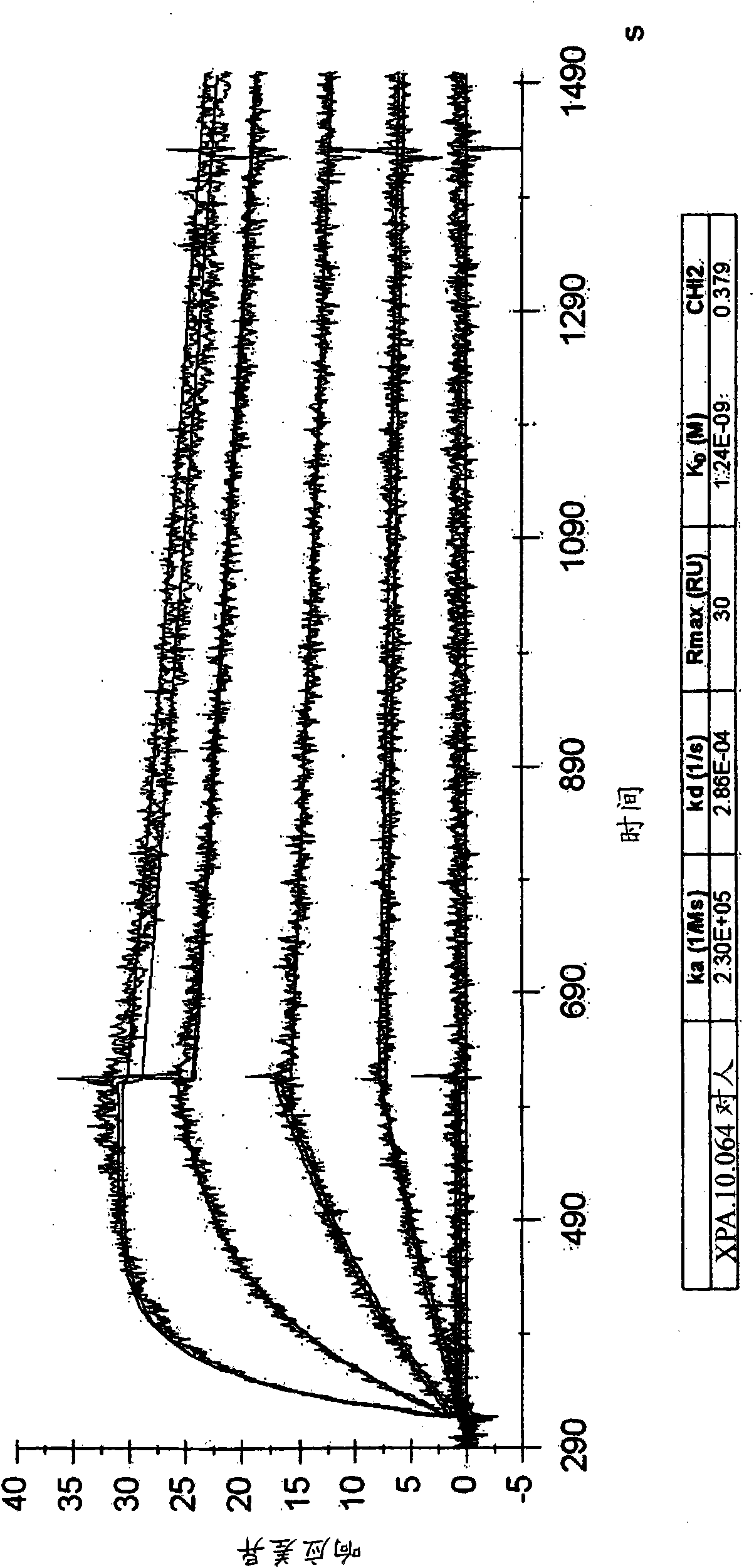
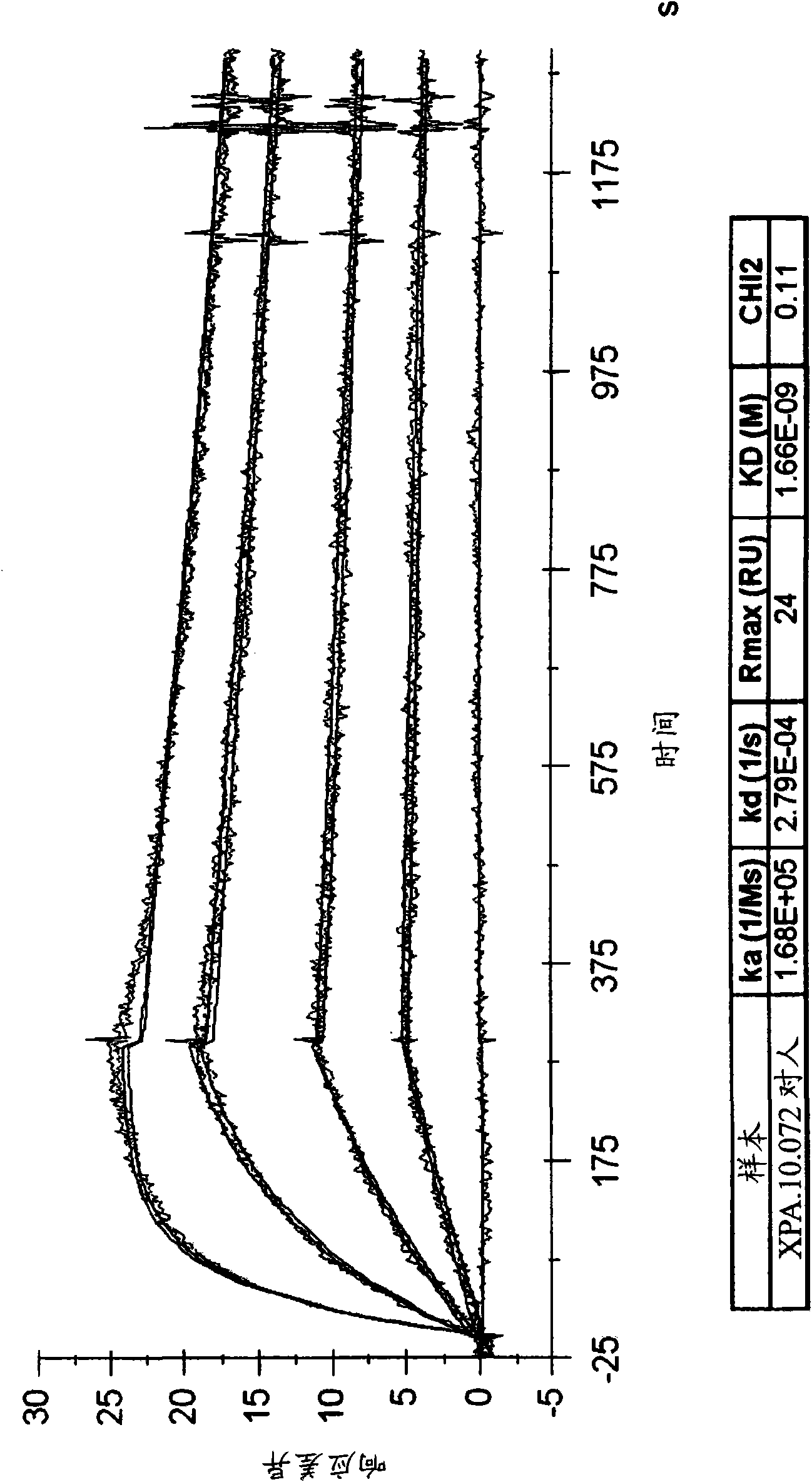
[0468] HVEGF 165 Two scFvs (XPA.10.064 and XPA.10.072) that inhibited binding to VEGF-R1 or VEGF-R2 by more than 60% were used for the conversion to scFv-Fc and / or IgG. figure 1 The heavy chain variable regions (including heavy chain CDRs) and light chain variable regions (including light chain CDRs) of XPA.10.064 and XPA.10.072 are shown. The heavy chain CDRs (e.g., HCDR1, HCDR2 and HCDR3) and light chain CDRs (for example, HCDR1, HCDR2 and HCDR3) are determined by the Kabat numbering system (Kabat, EA, et al. 1987, in Sequences of Proteins of Immunological Interest, US Department of Health and Human Services, NIH, USA) For example, LCDR1, LCDR2 and LCDR3). The HCDR1, HCDR2, and HCDR3 amino acid sequences of XPA.10.064 and XPA.10.072 are shown in SEQ ID NOs: 6, 7, and 8, respectively. The LCDR1, LCDR2, and LCDR3 amino acid sequences of XPA.10.064 are shown in SEQ ID NOs: 12, 13, and 14, respectively. The LCDR1, LCDR2, ...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com