Nucleic acid label for second-generation high-flux sequencing and design method thereof
A high-throughput, labeling technology, applied in the field of nucleic acid labeling, can solve the problems of reducing the misallocation rate, affecting the experimental results, high misallocation rate, etc., and achieving the effect of low cost
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
specific Embodiment
[0090] Specific examples are given below: Marking of 96 shellfish genome DNA samples
[0091] 1) Basic principles
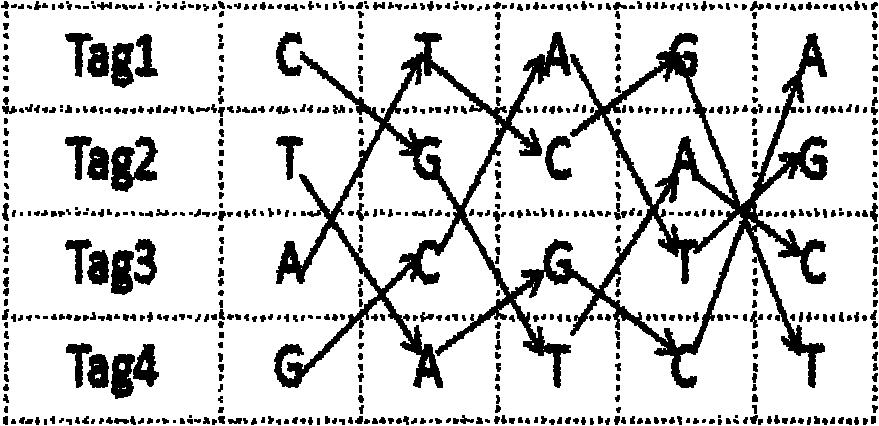
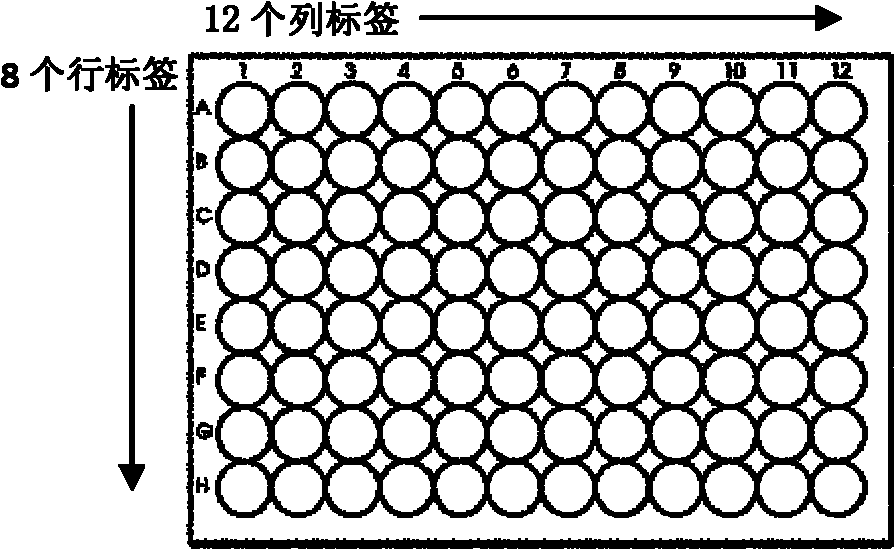
[0092] 96-well plates in a 12×8 format according to Molecular Biology Standards (see figure 2 ), the specific tags can also be designed as a combination of 12×8. Since tags can be attached to both ends of the DNA, 96 DNA samples can be labeled by the specific combination of 12×8 tags at both ends. Name the label that marks different rows as column-barcode (c-bar for short), 8 in total, and the samples in the same row of the 96-well plate mark the same row label; name the label that marks different columns as column label (row -barcode), referred to as r-bar, 12 in total, the samples in the same column of the 96-well plate are marked with the same column label (see figure 2 ).
[0093] use T c and T r Indicate the length of the c-bar and r-bar labels respectively; use p D Indicates the probability of assigning sequence errors to other samples, specific to...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com