Exploring method and exploring device for biological genome simple repeat sequence
A simple repetitive sequence and genome technology, applied in special data processing applications, instruments, electrical digital data processing, etc., can solve problems such as cumbersome and complex excavation processes
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment Construction
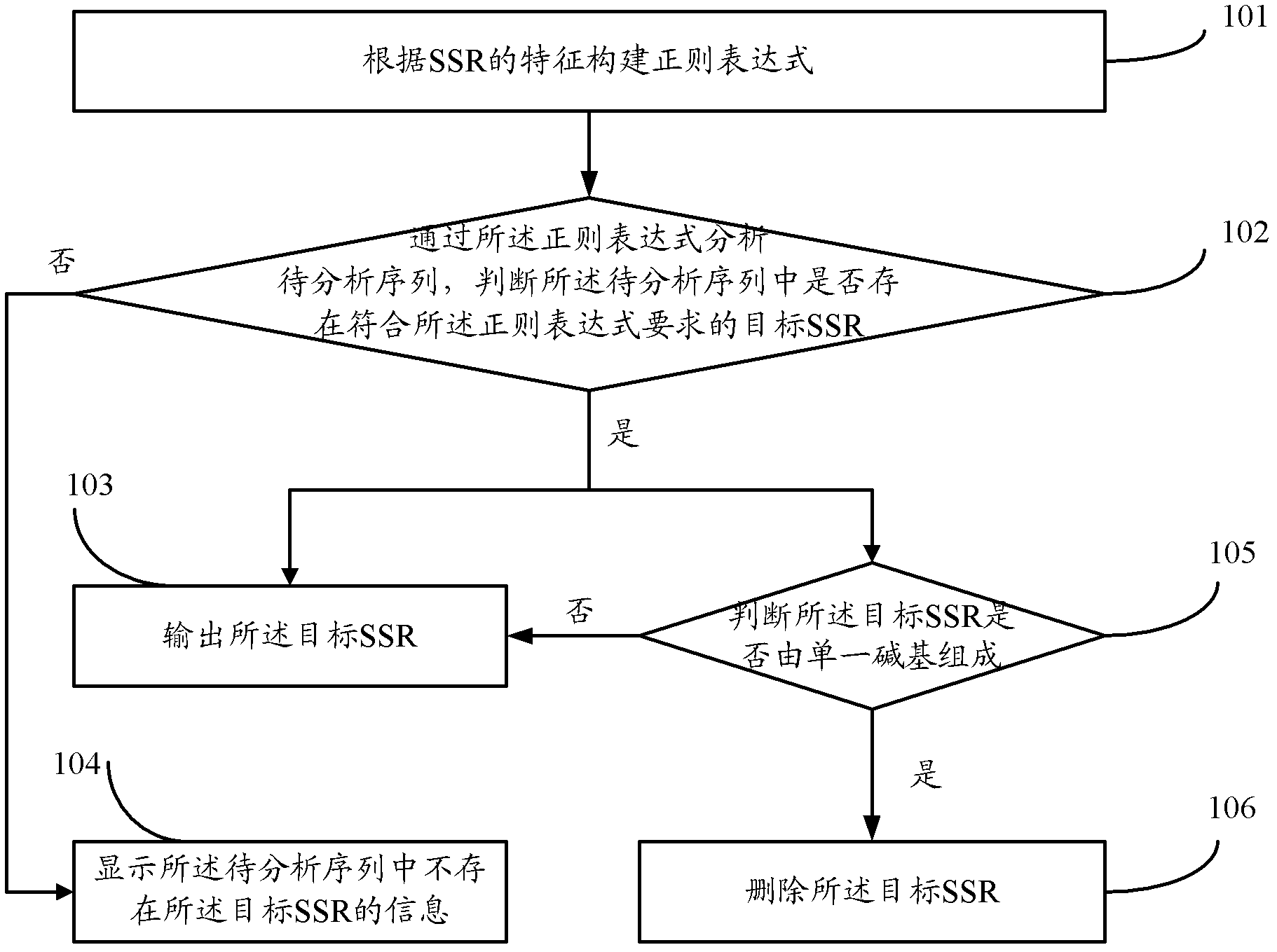
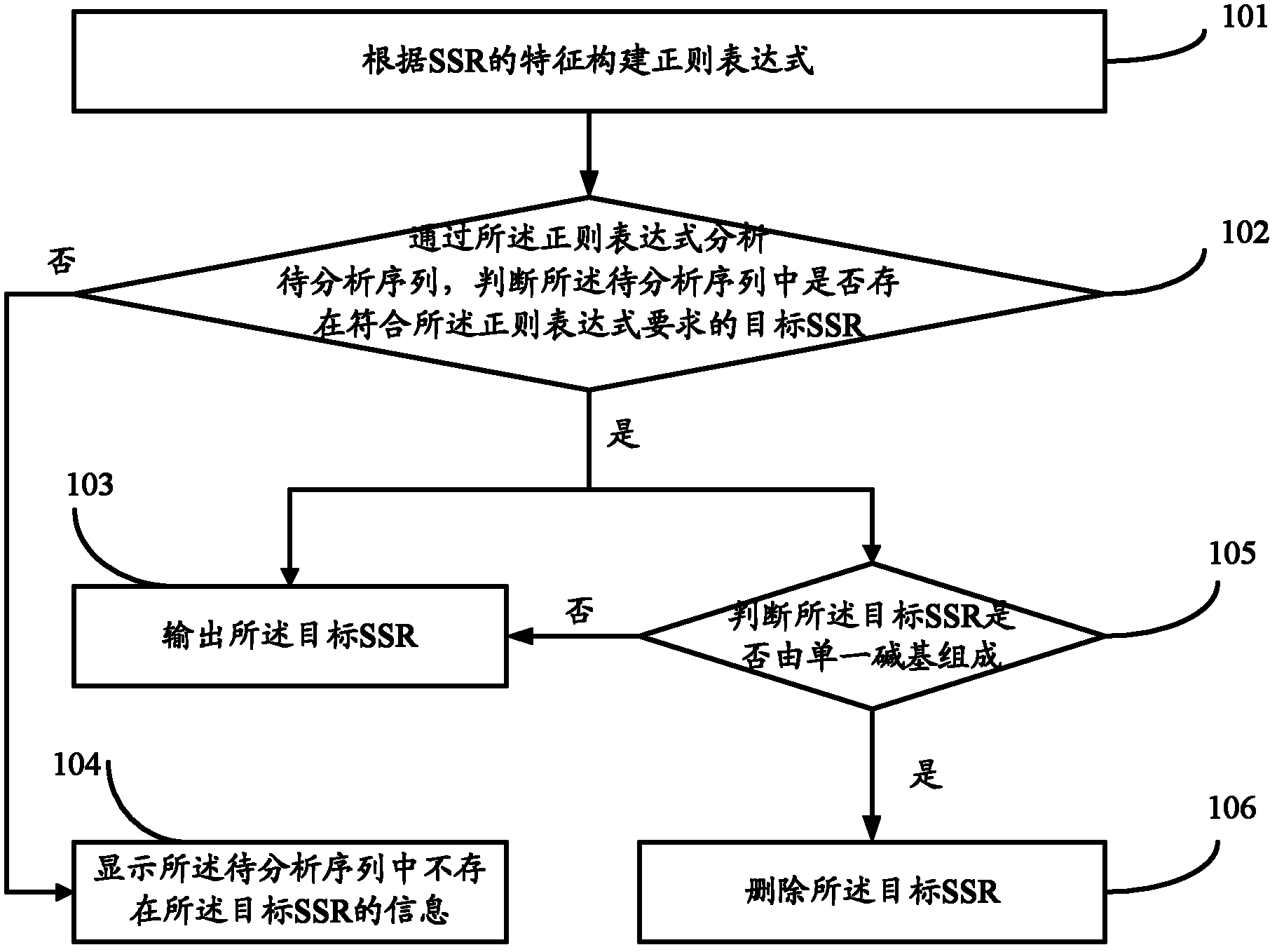
[0032] Such as figure 1 As shown, it is a schematic flow chart of the method for discovering the simple repeat sequence of the biological genome provided by the embodiment of the present invention, including the following steps:
[0033] Step 101, constructing a regular expression according to the characteristics of the simple repeat sequence SSR of the biological genome to be discovered;
[0034] Among them, the characteristics of the simple repeat sequence SSR of the biological genome that need to be discovered include:
[0035] The minimum length information of the motif in the simple repeat sequence SSR of the biological genome that needs to be discovered, the maximum length information of the motif, and the minimum number of repetitions information of the motif, wherein the motif refers to the motif in the SSR repeat unit.
[0036] The form of the regular expression constructed is: (.{i, j}?)(\1){k,}, wherein, i, j, k respectively represent the minimum length value of t...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com