Method for identifying varieties of Chinese cabbage by using composite expressed sequence tag-simple sequence repeat (EST-SSR) marker
A variety identification and Chinese cabbage technology, applied in the field of agricultural vegetable breeding and application, can solve problems such as restricting development and application, and achieve the effects of convenient operation, protection of crop varieties, and prevention of entering the market.
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
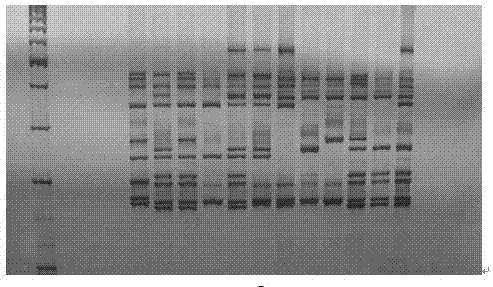
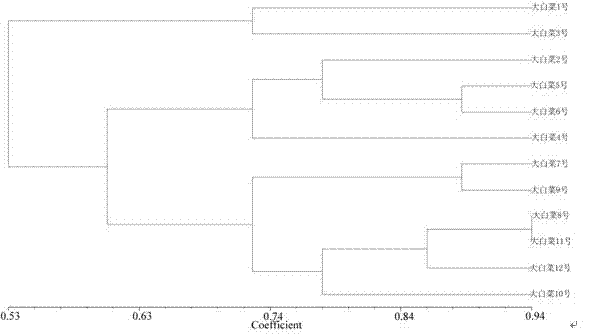
[0054] (1) The samples included 4 test Chinese cabbage hybrids No. 1 to No. 4, produced in Tianjin.
[0055] (2) Reagents: GoTaq? Master Mixes solution produced by Promega; Ⅰ or Ⅱ sets of composite EST-SSR primers synthesized by Shanghai Shenggong;
[0056] (3) The sequence table of EST-SSR primers of set I or set II is as follows:
[0057]
[0058] (4) Sampling and DNA extraction
[0059] Random samples were taken from farmers in the base. 10 plants of each variety were taken, 50-100 mg of tender leaves near the roots were taken and mixed as a sample, frozen ground with liquid nitrogen (Retsch MM400 bio-pulverizer), and CTAB lysis buffer (20 g / L CTAB, 1.4 M NaCl, 0.1 M Tris-HCl, 20 mM Na2EDTA) 500 μL, lysed in a constant temperature water bath (Neslab) at 65°C for 30 minutes, and subsequent DNA extraction and purification are performed according to the resin-type genomic DNA purification kit (Syberson). After DNA extraction, the concentration is uniformly diluted to 50±1 ng / μL, Nano...
Embodiment 2
[0067] (1) The samples included 4 test Chinese cabbage hybrids No. 5-8, produced in Tianjin.
[0068] (2) Reagents: GoTaq? Master Mixes solution produced by Promega; Ⅰ or Ⅱ sets of composite EST-SSR primers synthesized by Shanghai Shenggong;
[0069] (3) The sequence table of EST-SSR primers of set I or set II is as follows:
[0070]
[0071] (4) Sampling and DNA extraction
[0072] Random samples were taken from farmers in the base. 10 plants of each variety were taken, 50-100 mg of tender leaves near the roots were taken and mixed as a sample, frozen ground with liquid nitrogen (Retsch MM400 bio-pulverizer), and CTAB lysis buffer (20 g / L CTAB, 1.4 M NaCl, 0.1 M Tris-HCl, 20 mM Na2EDTA) 500 μL, lysed in a constant temperature water bath (Neslab) at 65°C for 30 minutes, and subsequent DNA extraction and purification are performed according to the resin-type genomic DNA purification kit (Syberson). After DNA extraction, the concentration is uniformly diluted to 50±1 ng / μL, Nanodrop ND...
Embodiment 3
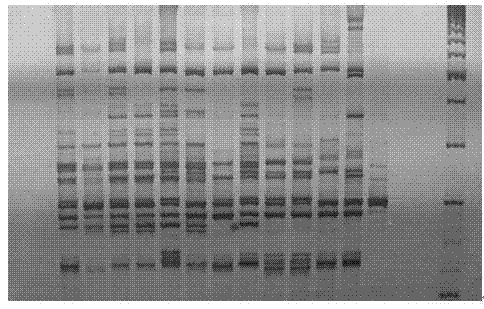
[0080] (1) The samples included 4 test Chinese cabbage varieties Hybrid No. 9-12, produced in Tianjin.
[0081] (2) Reagents: GoTaq? Master Mixes solution produced by Promega; Ⅰ or Ⅱ sets of composite EST-SSR primers synthesized by Shanghai Shenggong;
[0082] (3) The sequence table of EST-SSR primers of set I or set II is as follows:
[0083]
[0084] (4) Sampling and DNA extraction
[0085] Random samples were taken from farmers in the base. 10 plants of each variety were taken, 50-100 mg of tender leaves near the roots were taken and mixed as a sample, frozen ground with liquid nitrogen (Retsch MM400 bio-pulverizer), and CTAB lysis buffer (20 g / L CTAB, 1.4 M NaCl, 0.1 M Tris-HCl, 20 mM Na2EDTA) 500 μL, lysed in a constant temperature water bath (Neslab) at 65°C for 30 minutes, and subsequent DNA extraction and purification are performed according to the resin-type genomic DNA purification kit (Syberson). After DNA extraction, the concentration is uniformly diluted to 50±1 ng / μL, N...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com