Method for constructing sequencing library by cyclizing method
A sequencing library and cyclization technology, which is applied in the field of genetic engineering, can solve problems such as the inability to construct small fragment nucleic acid molecular sequencing libraries, and achieve the effects of expanding the scope of application and improving efficiency
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
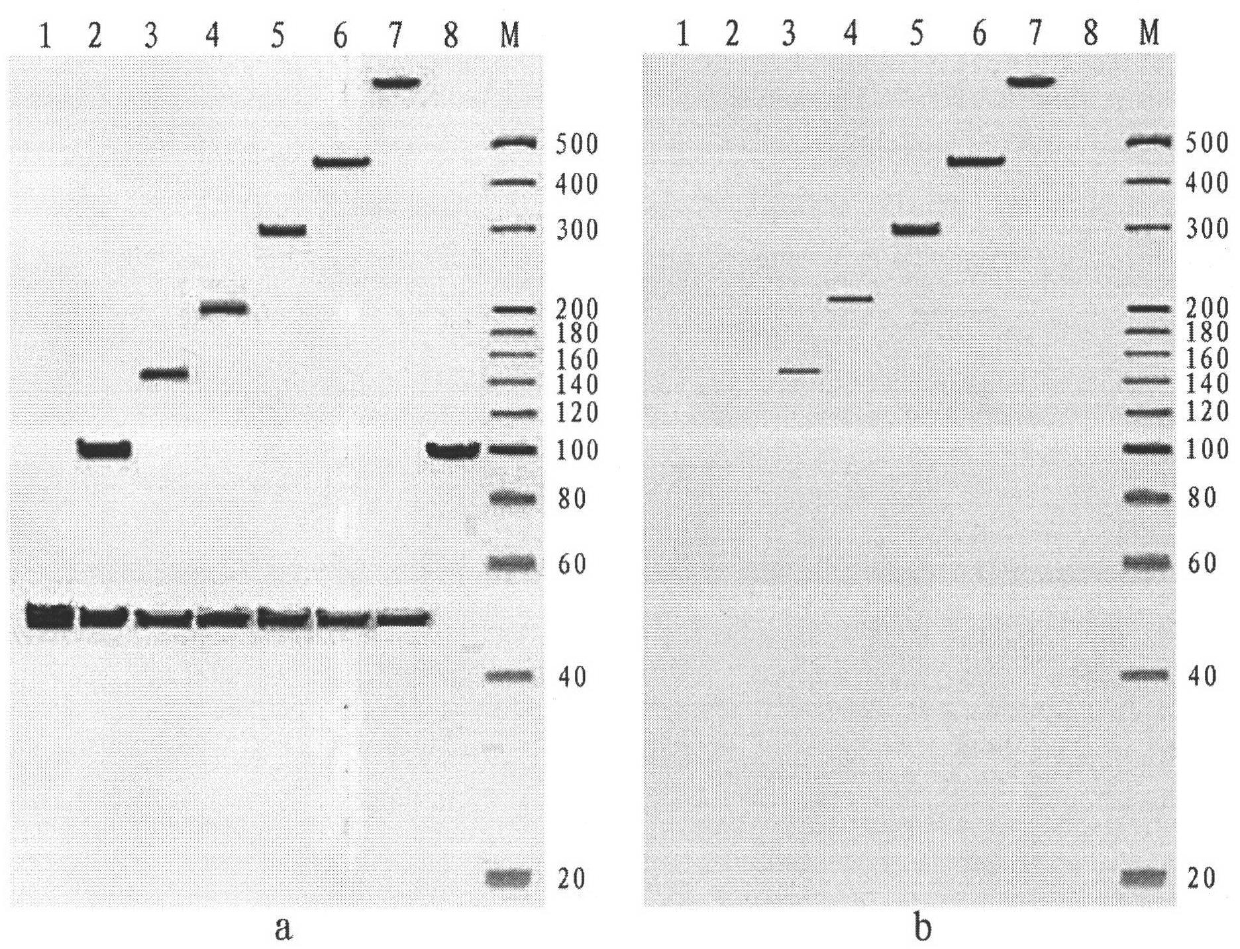
Image
Examples
Embodiment approach
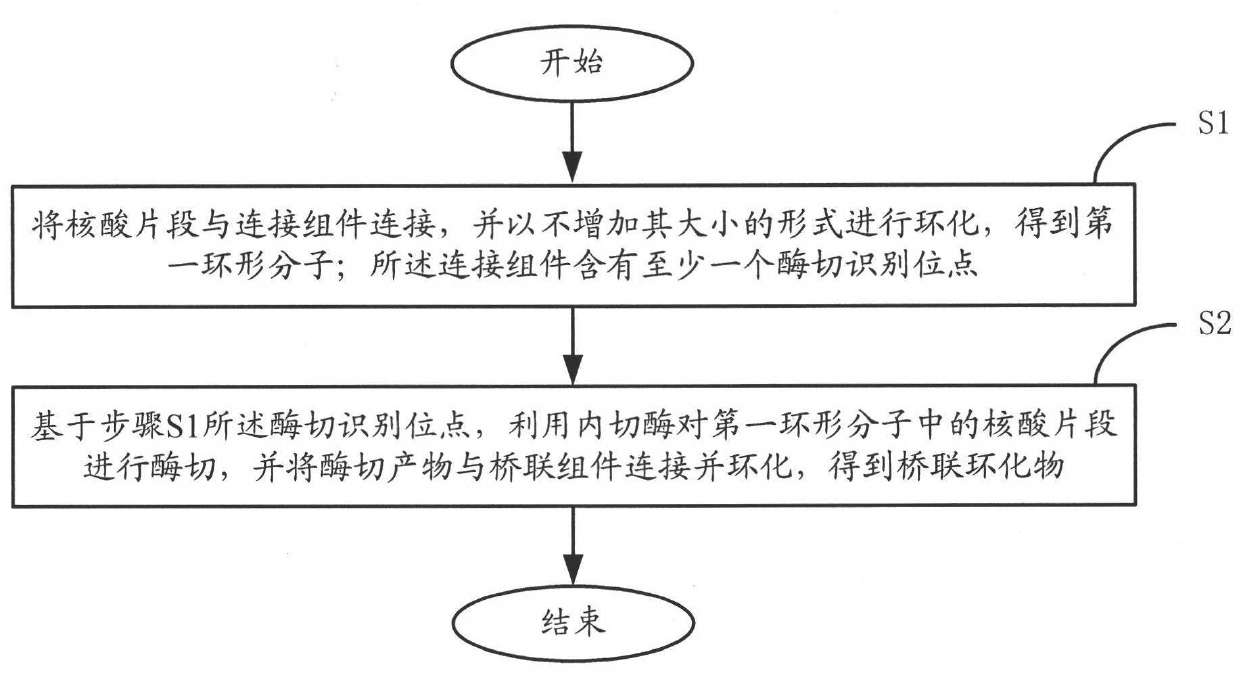
[0054] figure 1 An embodiment of the method for constructing a sequencing library of the present invention is shown, comprising the steps of:
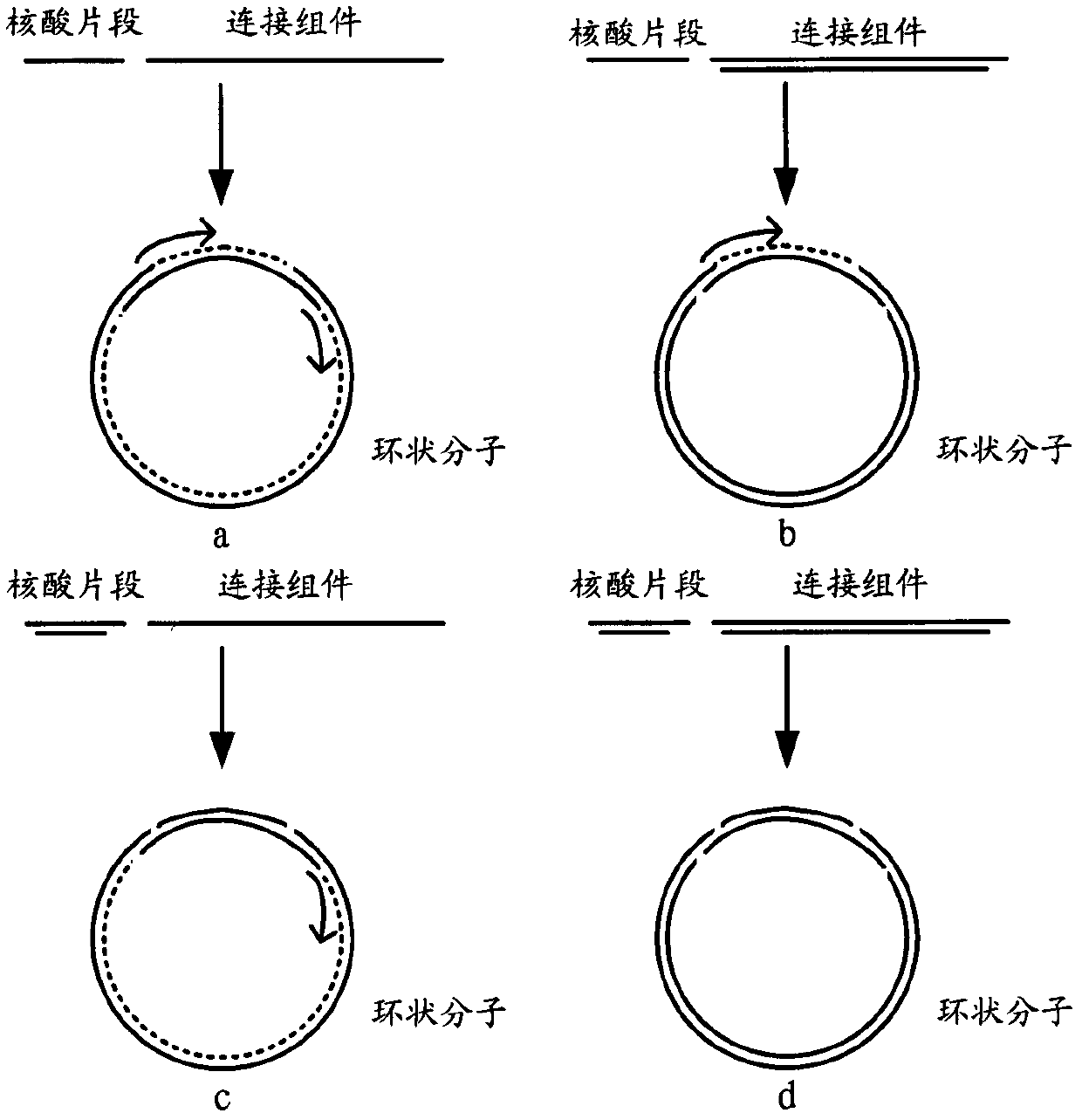
[0055]S1. Ligating the nucleic acid fragments with the connecting element, and performing circularization in a form that does not increase its size, to obtain a first circular molecule; the connecting element contains at least one restriction enzyme recognition site;
[0056] S2. Based on the digestion recognition site described in step S1, the nucleic acid fragment in the first circular molecule is digested with an endonuclease, and the digested product is connected to the bridging component and cyclized to obtain a bridging cyclized product.
[0057] It should be noted:
[0058] In step S1, the nucleic acid fragment can be any fragment in the genome, including but not limited to a gene, a part of a gene, a regulatory sequence, an intron or a part of an intron; it can also be genomic DNA, cDNA, RNA ( Including but not limited to mRN...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com