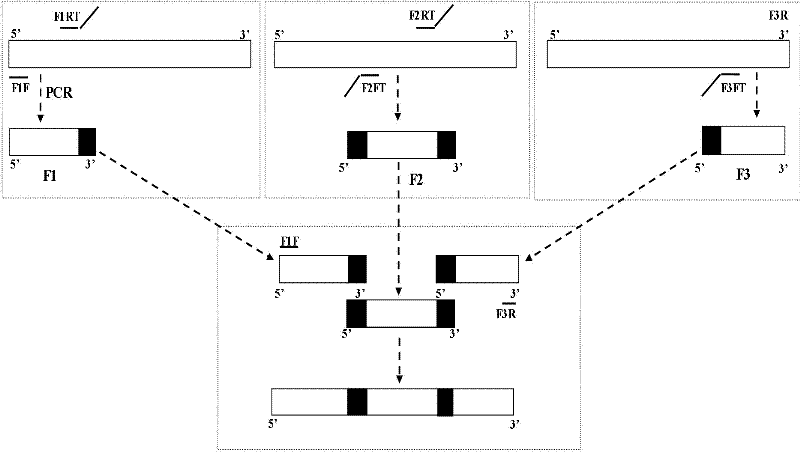
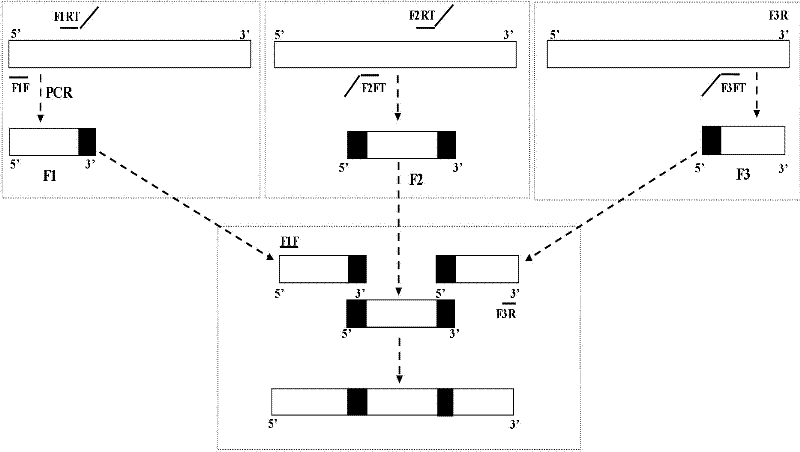
Rapid bidirectional multilocus gene mutation method
A multi-site mutation and multi-site technology, applied in the field of bioengineering, can solve the problems of long time and limited mutation sites.
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
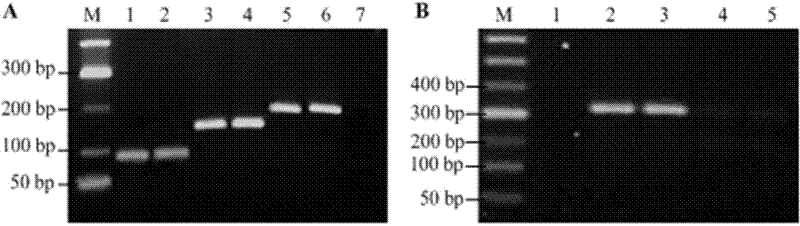
[0012] Example 1: Obtaining a 324 bp DNA fragment containing two mutations
[0013] 1. Design of primers
[0014] According to the quasi-mutant site and its sequence characteristics of the selected DNA fragment (see sequence table SEQ ID No. 1), three pairs of specific primers were designed to amplify the selected DNA fragment and its adjacent upstream and downstream fragments ( F1: upstream fragment, F2: target sequence, F3: downstream fragment). The designed primer sequences are shown in Table 1, where the underlined part is the extended sequence, that is, the overlapping part.
[0015] Table 1 Primer sequences
[0016] name Primer sequence (5'-3') Length (nt) F1F AGATCTGAGCCTGGGAG 17 F1RT GGTTCCGTTATTTCCCAAGTT TTAAGCAGTGGGTTCCCTAG 40 F2FT AACTTGGGAATAACGGAACC GAGTGCTTCACTTGATACAG 40 F2RT GCCAATCTCTCAATCGCGGC ATTTTCCACACTGACTAAAAG 41 F3FT GCCGCGATTGAGAGATTGGC CAGGGACCTGAAAGCGAAAG 40 F3R CACCCATCTC...
Embodiment 2
[0023] Embodiment 2: the length is 324 bp, contains the acquisition of two mutant DNA fragments
[0024] In this example, except for the template and primers, the two rounds of PCR amplification conditions are the same as in Example 1. The final product is also 324bp in length and contains a point mutation sequence at both ends. The primers are shown in Table 2, and the underlined part indicates the overlapping part. For the mutation product, see SEQ. ID. No.13 in the sequence listing.
[0025] Table 2 Primer sequences
[0026] name Primer sequence (5'-3') Length (nt) F1F AGATCTGAGCCTGGGAG 17 F1RT 4m GGTACCGTTATTTCCCAAGTT TTAAGCAGTGGGTTCCCTAG 40 F2FT 4m AACTTGGGAATAACGGTACC GAGTGCTTCACTTGATACAG 40 F2RT 4m GCCAATCTCTCCAATCCCGGC ATTTTCCACACTGACTAAAAGG 42 F3FT 4m GCCGGGATTGAGAGATTGGC CAGGGACCTGAAAGCGAAAG 40 F3R CACCCATCTCTCTCCTTCTA 20
[0027] Wang Shuqi
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com