Optimized endonucleases and uses thereof
一种内切核酸酶、多核苷酸的技术,应用在优化的内切核酸酶领域,能够解决非意图性脱靶作用、减少切割活性等问题
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0326] Example 1: Construction of a sequence-specific DNA endonuclease expression cassette for expression in E. coli
Embodiment 1a
[0327] Example 1a: Basic construct
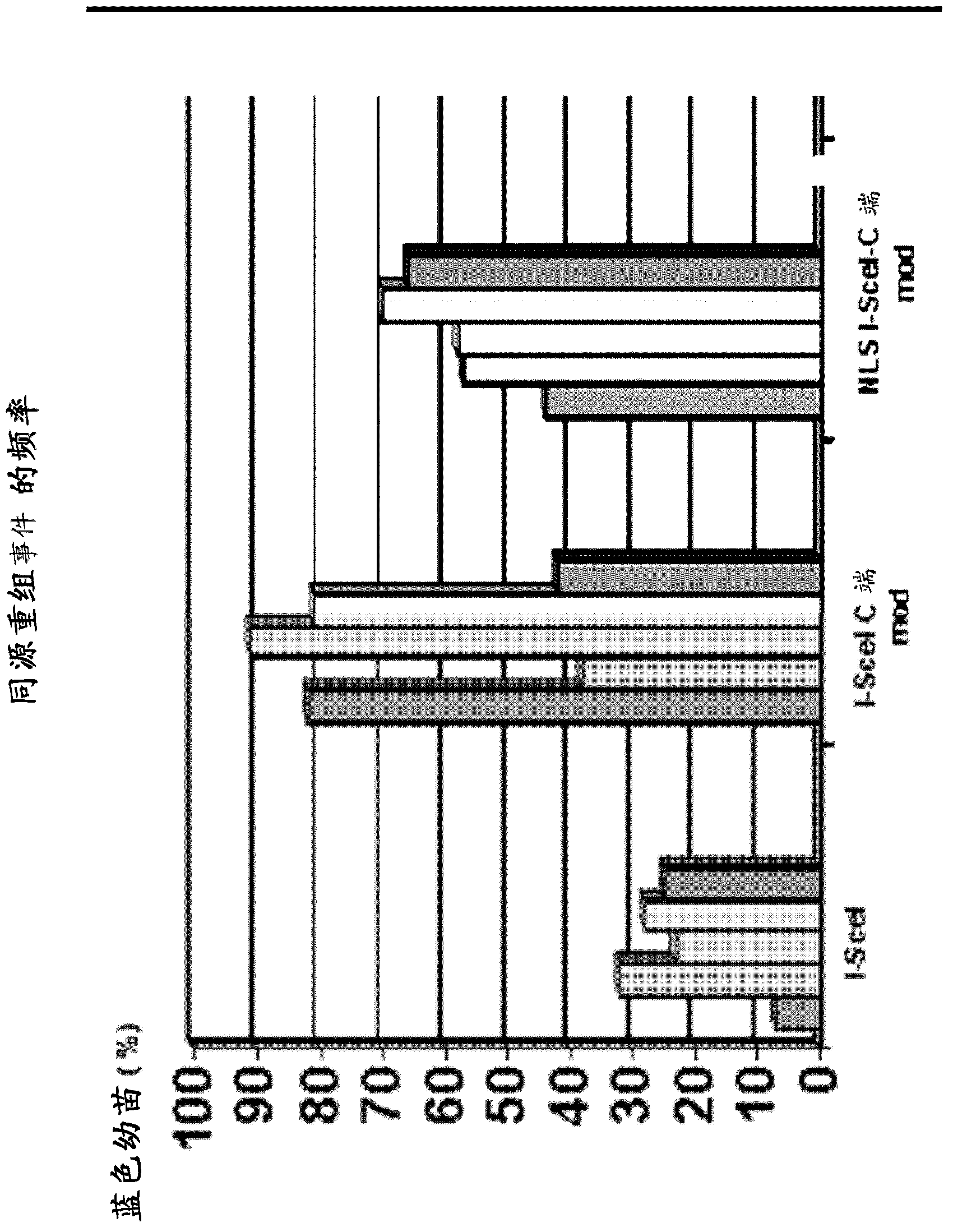
[0328] In this example, we show the general outline of the vector named "Construct I", which is suitable for transformation in E. coli. This general profile of the vector contains an ampicillin resistance gene for selection, an origin of replication for E. coli, and the gene araC (which encodes an arabinose-inducible transcriptional regulator). SEQ ID NO: 7 shows the stretch of "NNNNNNNNNN". This represents a placeholder for genes encoding different versions of sequence-specific DNA endonucleases. Different genes can be expressed from the arabinose-inducible pBAD promoter (Guzman et al., J Bacteriol 177:4121-4130 (1995)), and the gene sequences encoding different nuclease versions are given in the following examples.
[0329] The control construct in which the placeholder is replaced by the sequence of I-Scel (SEQ ID NO: 8) is called VC-SAH40-4.
Embodiment 2
[0330] Example 2: Escherichia coli plasmid encoding a stable version of nuclease
[0331] Different destabilizing sequences can be identified in the amino acid sequence of I-Scel. Among them are the weak C-terminal sequence of PEST containing amino acid residues 228 to 236, and the N-terminal sequence that shows similarity to the KEN motif (Pfleger and Kirschner, Genes and Dev. 14: 655-665 (2000)). According to the N-terminal rule, the second amino acid residue of I-Scel confers protein instability.
[0332] In order to test the influence of these sequences on the stability of nucleases, different versions of I-SceI were generated by PCR, which deleted the N-terminal amino acid, the C-terminal 9 amino acids, or both. These constructs are expressed according to "Construct I" described in Example 1a). Therefore, the placeholders are replaced by multiple sequences (shown in SEQ ID NO: 2, 3, 5) encoding each version of the nuclease. The plasmids are called VC-SAH43-8 (I-SceI with sh...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com