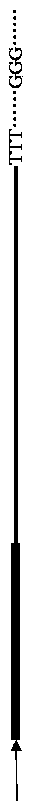Chromosome walking method based on PCR (Polymerase Chain Reaction) technology
A technology of chromosome walking and technology, applied in the field of chromosome walking, can solve the problems of limited randomness, low specificity, and limited template binding force of ligation-mediated methods, and achieve the elimination of enzyme digestion and ligation steps, broad The effect of application prospect and long walking distance
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment Construction
[0018] In the following, a specific cell is taken as an example to describe the embodiment of the present invention in detail.
[0019] 1. Preparation of eukaryotic cell genome transfected with exogenous DNA: co-transfect goat fetal fibroblasts with pIRES2-EGFP-attB-OTLR9 and pCMVInt, the φC31 integrase expressed by pCMVInt can mediate pIRES2-EGFP-attB-OTLR9 The vector is site-specifically integrated into the cell genome. After 24 hours of transfection, the cells were screened with G418. After positive cell clones appeared, the positive cell monoclonal expansion culture was continued, and then the cells were collected, and their genomes were extracted using a cell genome extraction kit.
[0020] 2. Amplification of the target DNA single strand: design two nested specific primers P1 5'-CGTATGTTCCCATAGTAACGCCAAT extending to the unknown sequence end at the right CMV promoter region of the pIRES2-EGFP-attB-OTLR9 vector integration site attB -3' and P2 5'-TACATGACCTTATGGGACTTTCCT...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com