Method for constructing gene site-directed mutation
A technology of gene site-directed mutation and construction method, which is applied in the field of gene site-directed mutation construction in rat embryonic cells. Experimental cost, effect of high proportion of mutants
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Benefits of technology
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0075] Example 1 Construction of Mc4r Gene Knockout Rats by Injecting RNA in Rat Cells
[0076] 1. Construction of NLS-hCas9-NLS in vitro transcription vector
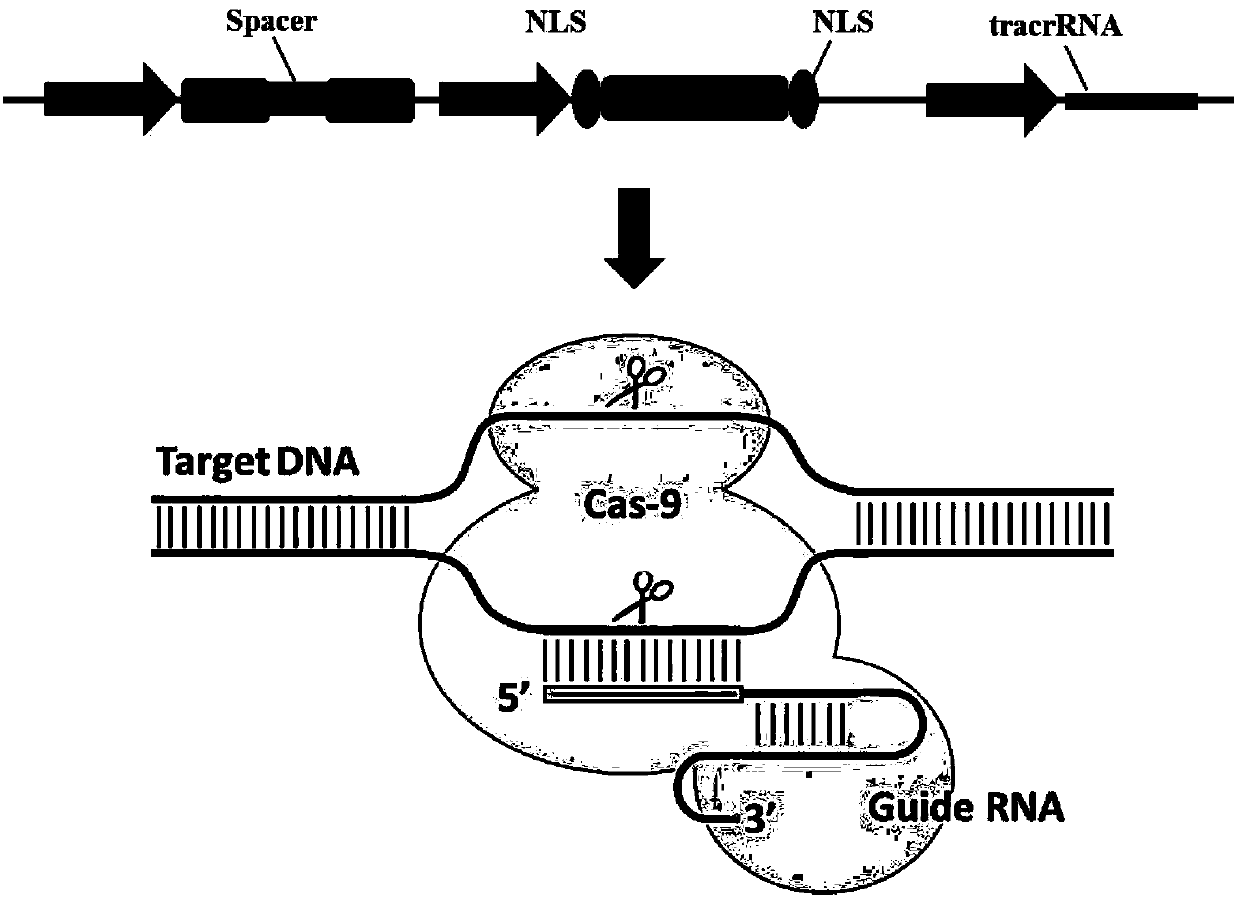
[0077] Such as figure 2 As shown, the Cas9 protein expression module consists of the SP6 promoter sequence from the 5' end to the 3' end, the N-terminal nuclear localization signal (Nuclear localization sequence, NLS), the humanized Cas9 coding DNA sequence, the C-terminal nuclear localization signal, Polyadenylic acid (polyA) composition. The specific construction strategy is, on the basis of the vector pX260 (Addgene plasmid #42229), introduce the SP6 promoter and the Kozak sequence through the NcoI restriction endonuclease site overlapping with the start codon of the NLS-hCas9-NLS coding sequence. That is, synthesize single-stranded oligonucleotides P1 (SEQ ID NO.1) and P2 (SEQ ID NO.2), use PNK phosphatase to add 5' phosphate groups to the above-mentioned single-stranded oligonucleotides, and anneal them Ligate...
Embodiment 2
[0102] Example 2 Construction of Mc3r / Mc4r knockout rats by injecting CAS protein and gRNA in rat cells
[0103] 1. Construction of CAS nuclease prokaryotic expression vector PET-28a-H6-3FLAG-NLS-CAS9-NLS
[0104] Such as Image 6 As shown, the prokaryotic expression vector of Cas9 fusion protein consists of T7 promoter sequence, His6Tag, 3×Flag, N-terminal nuclear localization sequence (NLS1), and humanized Cas9 encoding DNA from the 5' end to the 3' end. Sequence, C-terminal nuclear localization signal NLS2 constitutes. First, based on the NLS-hCas9-NLS in vitro transcription vector, His6 and 3×FLAG tags, as well as a new N-terminal NcoI restriction enzyme site and a C-terminal EcoRI restriction enzyme site were introduced by overlapping PCR. A new H6-3FLAG-NLS-CAS9-NLS junction was inserted through the NcoI and EcoRI restriction sites on PET-28a and used as translation initiation for protein expression through the start codon contained in the NcoI restriction site locati...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com