Method for obtaining oligonucleotide probe
An oligonucleotide probe and oligonucleotide technology, applied in the field of plant molecular chromosome engineering breeding, can solve problems such as high cost, complicated probe labeling technology, and probe labeling failure, so as to improve efficiency and avoid probe The effect of labeling failure and experimental cost reduction
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment Construction
[0038] In order to make the object, technical solution and advantages of the present invention clearer, the present invention will be further described in detail below in conjunction with the examples. It should be understood that the specific embodiments described here are only used to explain the present invention, not to limit the present invention.
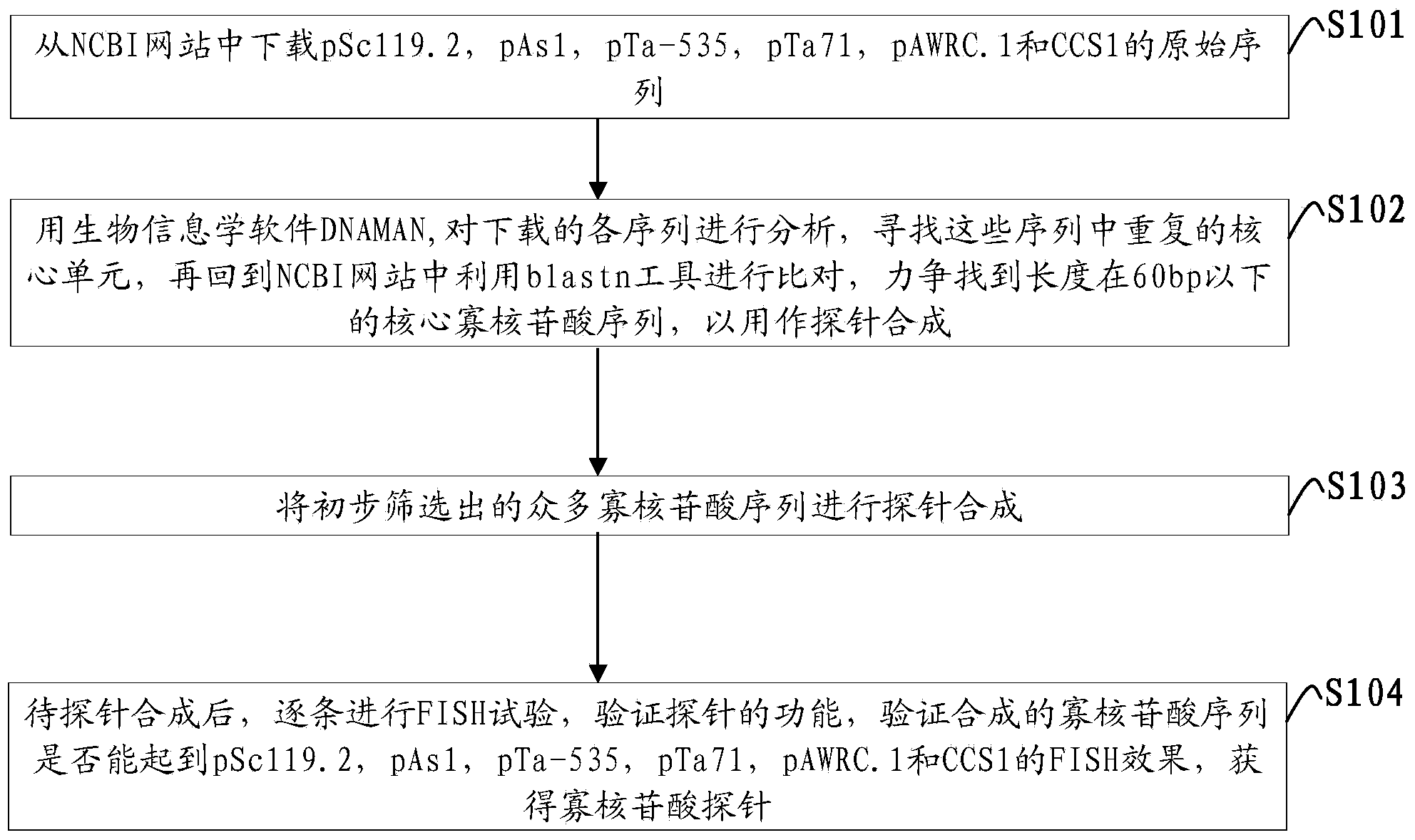
[0039] Such as figure 1 Shown, the method for obtaining oligonucleotide probe of the embodiment of the present invention comprises the following steps:
[0040] S101: Download the original sequences of pSc119.2, pAs1, pTa-535, pTa71, pAWRC.1 and CCS1 from the NCBI website;
[0041]S102: Use the bioinformatics software DNAMAN to analyze the downloaded sequences, find the repeated core units in these sequences, and then return to the NCBI website and use the blastn tool for comparison, and strive to find core oligonucleotides with a length of less than 60 bp Acid sequences for probe synthesis;
[0042] S103: Synthesizing the ...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com