Delta12-fatty acid dehydrogenase gene and application of gene
A fatty acid dehydrogenase and unsaturated fatty acid technology, applied in genetic engineering and biological fields, can solve problems such as difficult separation and difficult biochemical characteristics, and achieve the effects of improving oil quality, low cost and short production cycle
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0023] Example 1: Isolation of Δ from Mortierella chrysogenum M6-22 12 - Nucleotide sequence of fatty acid dehydrogenase
[0024] Genomic DNA of Mortierella flavinum M6-22 was extracted using the Ezup Column Fungal Genomic DNA Extraction Kit (product number: SK8259) of Sangon Bioengineering (Shanghai) Co., Ltd., and 1ul was used as a template for polymerase chain reaction. response, according to the published Δ 12 -Degenerate primers (primer 1 and primer 2) were designed for the amino acid sequence of the histidine conserved region Ⅰ and Ⅲ region of the fatty acid dehydrogenase homologous sequence, and PCR amplification was carried out on a PCR machine (BIOER company) with the above-mentioned extracted DNA template , the primers, components and amplification conditions used in the reaction are as follows:
[0025] Primer 1: FD12CRF1: 5`-CA(T / C)GA(A / G)TG(T / C)GGICA(T / C)CA(A / G)(G / T)CITT-3`
[0026] Primer 2: FD12CRR3: 5`-(A / T)A(A / G)TG(A / G)TGI(A / G)(A / G / C)IAC(A / G)TGIGT-3`
[00...
Embodiment 2
[0048] Example 2: Homology Search of Isolated Nucleotide Sequences
[0049] will infer that Δ 12 - The amino acid sequence of fatty acid dehydrogenase was searched for homology on Genbank, and most of the obtained homologous sequences encoded Δ 12 -Sequences of fatty acid dehydrogenases, a few are hypothetical protein sequences, select Δ that has been functionally verified 12 - Fatty acid dehydrogenase for homology comparison, which is compared with Mucor circinosa ( Mucor circinelloides )Δ 12 - Fatty acid dehydrogenase has the highest homology, with an identity of 58% ( figure 1 ). figure 1 Shows the separation of Δ in Mortierella flavinum M6-22 of the present invention 12 - Fatty acid dehydrogenase and Mucor circinatus Δ 12 -Comparison of amino acid sequence homology of fatty acid dehydrogenase (GenBank: BAB69056.1), MID12 is Mortierella chrysogenum M6-22Δ of the present invention 12 - Fatty acid dehydrogenase, MCD12 for Mucor circinatus Δ 12 - Fatty acid dehydrog...
Embodiment 3
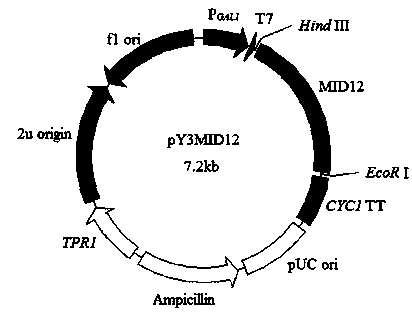
[0050] Example 3: Construction of Saccharomyces cerevisiae recombinant expression vector
[0051] According to the sequence of the coding region shown in SEQ ID NO:1, a pair of gene-specific amplification primers (primer 3 and primer 4) were designed to isolate its potential open reading frame sequence:
[0052] Primer 3: MID12F:5`-ATT AAGCTT ATGCCACCCAACGTAACGTCT-3`
[0053] Primer 4: MID12R:5`-GCA CTCGAG TTAGTTTTTTGAAGAACAAGACGT-3`
[0054] The underlined parts at the 5‵ ends of these two primers respectively contain Hind III and xho Ⅰ Restriction site, the amplification conditions and reaction components used are the same as the PCR reaction using cDNA as template, and the sequencing result of the amplified product shows that it is consistent with the sequence of 1-1194bp shown in SEQ ID NO:1. Then take 25ul of PCR products and 10ulpYES3 / CT for double enzyme digestion, recover the large fragments, and use T4 ligase to ligate at 16 degrees for 4 hours. The ligated p...
PUM
 Login to view more
Login to view more Abstract
Description
Claims
Application Information
 Login to view more
Login to view more - R&D Engineer
- R&D Manager
- IP Professional
- Industry Leading Data Capabilities
- Powerful AI technology
- Patent DNA Extraction
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic.
© 2024 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap