A gene chip for detecting 11 common infectious diarrhea pathogens and its application
An infectious diarrhea and gene chip technology, applied in the field of gene chips, can solve the problems of high similarity and inability to design probes, and achieve the effects of strong specificity, good sensitivity and high detection throughput
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0054] Embodiment 1 gene chip detection probe
[0055] 1. PCR amplification and electrophoresis detection
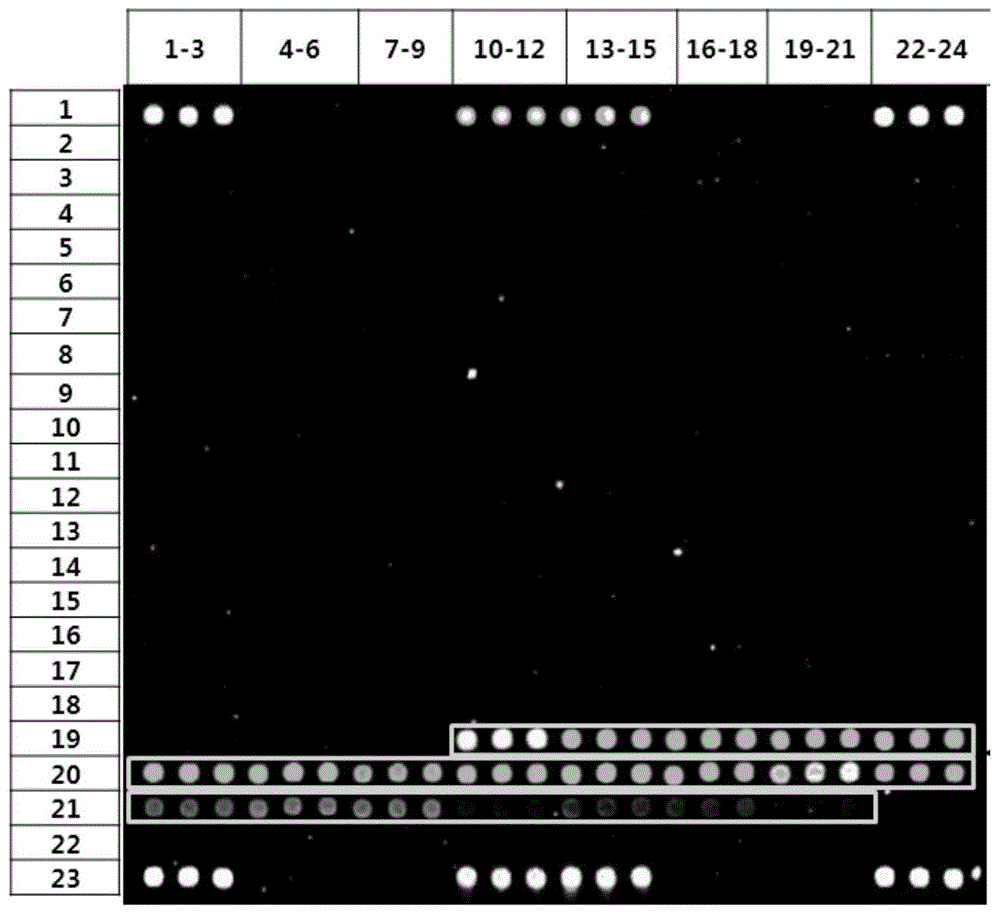
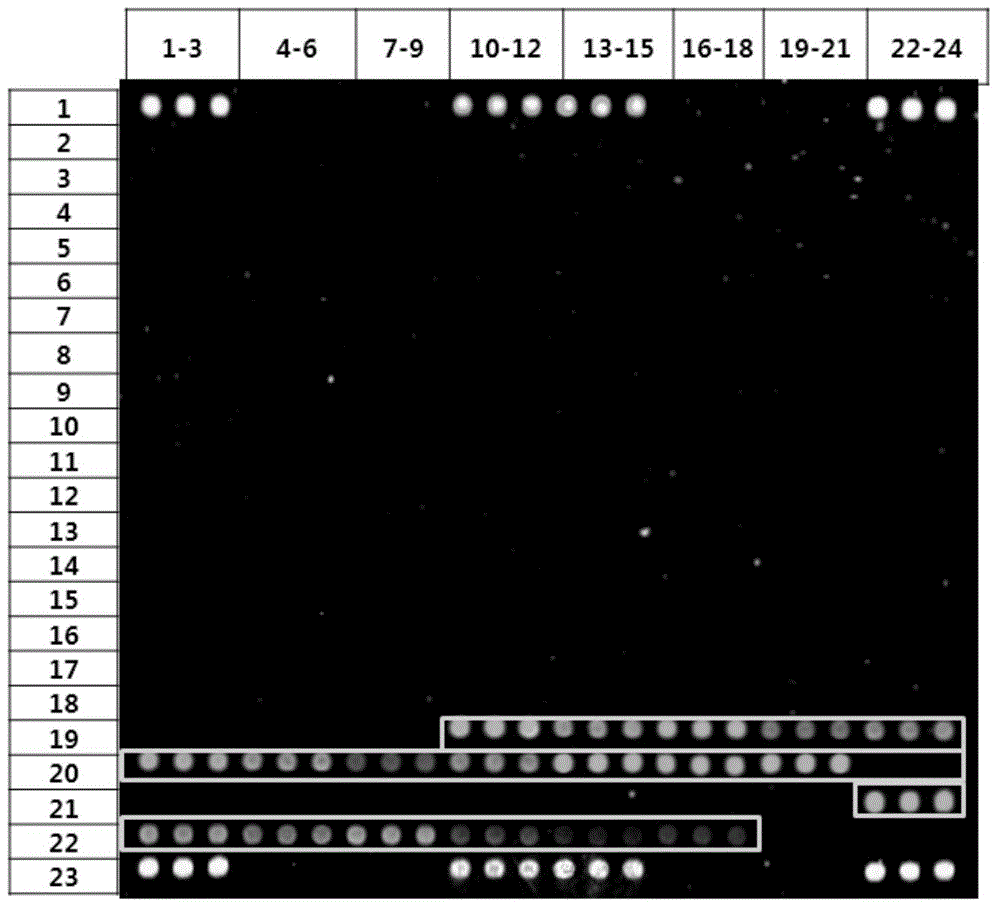
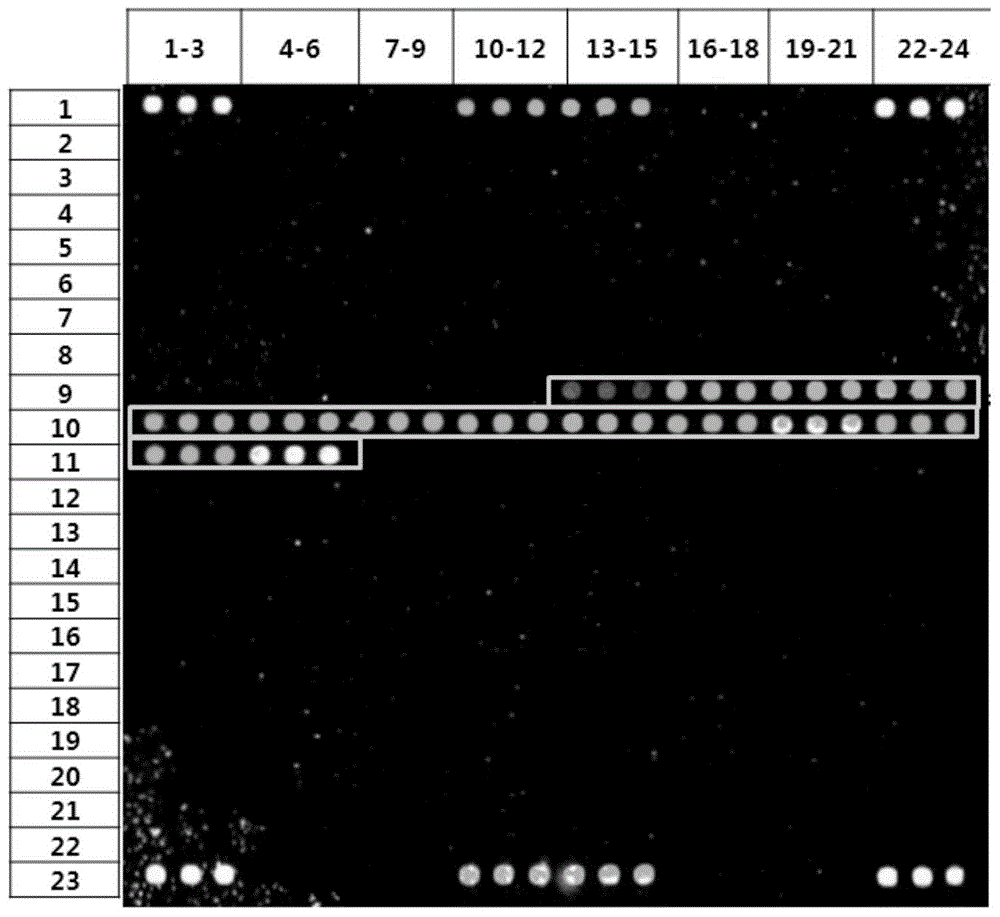
[0056] Carry out PCR amplification and electrophoresis detection to the 16SrRNA gene of sample bacteria (Vibrio vulnificus, Shigella dysenteriae, Escherichia coli O157:H7, Salmonella, see table 1.1), primer sequence such as SEQIDNO:118-119 As shown, the amplified electrophoresis results are as follows Figure 14 As shown, the sequencing results of the amplified products are shown in Table 1.2, and some screenshots of the sequencing peaks are shown in Figure 13 shown.
[0057] Table 1. Annotation of electropherogram after 116SrRNA gene PCR
[0058]
[0059] Table 1.2 Sequencing test results
[0060]
[0061] 2. Probe Design
[0062] For the microorganisms provided in the diarrhea project, a probe with a length of 40mer was designed to distinguish the target microorganisms. The microorganisms provided in the project are shown in Table 2.1 below:
[0063] Table...
Embodiment 2
[0079] Example 2 gene chip
[0080] The chip manufacturing process of the diarrhea pathogenic bacteria gene chip includes: preparation of spotting probes, preparation of aldehyde-based substrates, chip printing, fixing and scanning, chip packaging and other steps. For the process route, see Figure 15 .
[0081] 1. Preparation of sampling probe
[0082] Crystal core Gene chip spotting solution and water, prepare the oligonucleotide probe into a specific concentration spotting solution, and then transfer it to the corresponding position of the 384-well plate according to the dot array pattern, and seal it with parafilm, ready for spotting.
[0083] 1.1 Selection of sampling solution
[0084] The crystal core used in the preparation of the gene chip of diarrhea pathogenic bacteria The gene chip spotting solution is a product independently developed and commercialized by Boao Biotechnology Co., Ltd. (catalogue number: 440010). Comparison of the results after spotting with...
Embodiment 3
[0103] Embodiment 3 uses gene chip to carry out sample detection
[0104] 1. Sample collection and processing
[0105] The pathogenic microorganisms detected were enterohaemorrhagic Escherichia coli (E.coli), Shigella dysenteriae (S.dysenteriae), Salmonella (Salmonella), Vibrio vulnificus (V.vulnificus), Vibrio parahaemolyticus (V. .parahaemolyticus), Vibrio cholerae (V.cholerae), Proteus vulgaris (P.vulgaris), Vibrio alginolyticus (V.alginolyticus), Vibrio furnissii (V.furnissii), Aeromonas hydrophila (A.hydrophila), Campylobacter jejuni (Campylobacter); the DNA of the sample was extracted from the above-mentioned microbial samples by a bacterial genome nucleic acid extraction kit (purchased from QIAGEN).
[0106] 2. Amplification and labeling of sample nucleic acids
[0107] 2.1 Polymerase chain reaction
[0108] 2.1.1 Universal amplification primers
[0109] Design and screen a universal primer pair that can amplify the full length of the target gene as broadly as possibl...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com