RNA interferases and methods of use thereof
A technology of use and enzyme polypeptide, applied in the direction of DNA preparation, recombinant DNA technology, biochemical equipment and methods, etc.
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment I
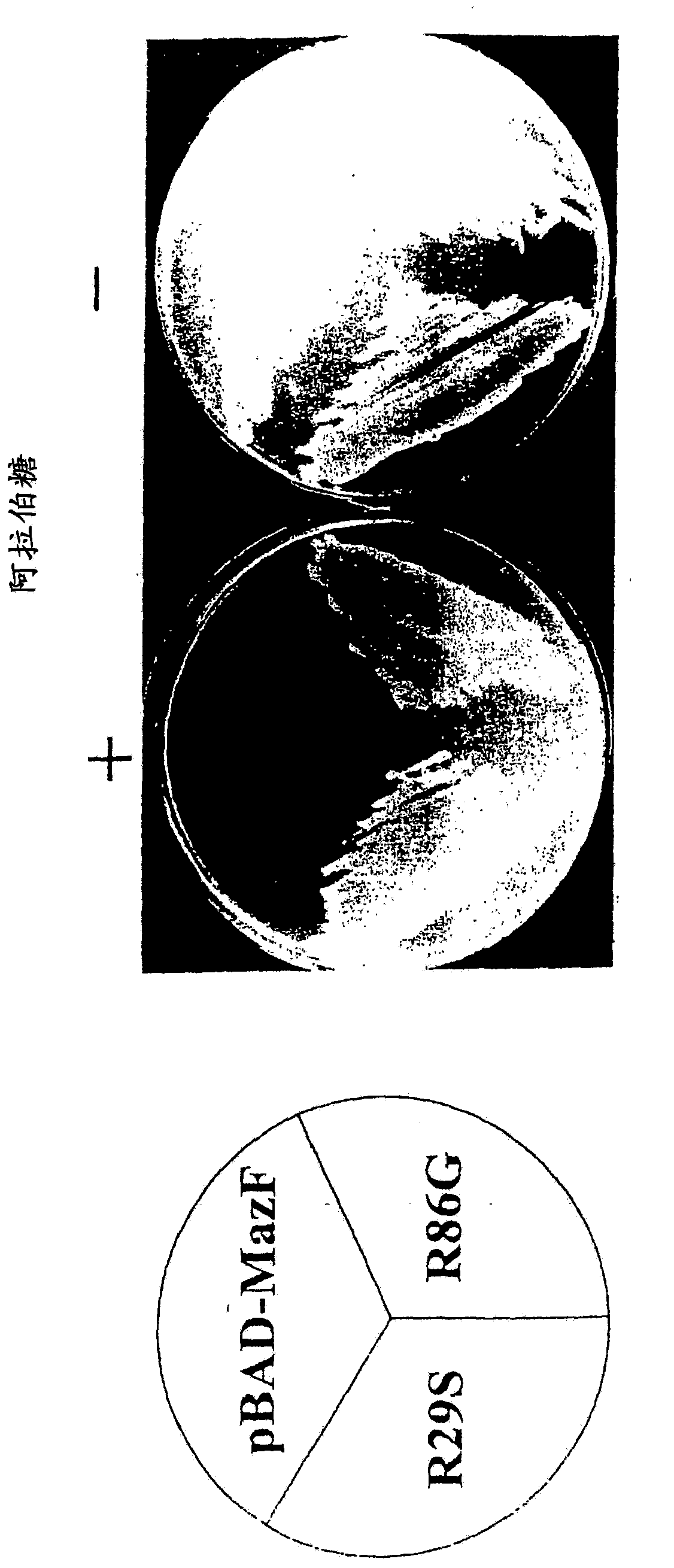
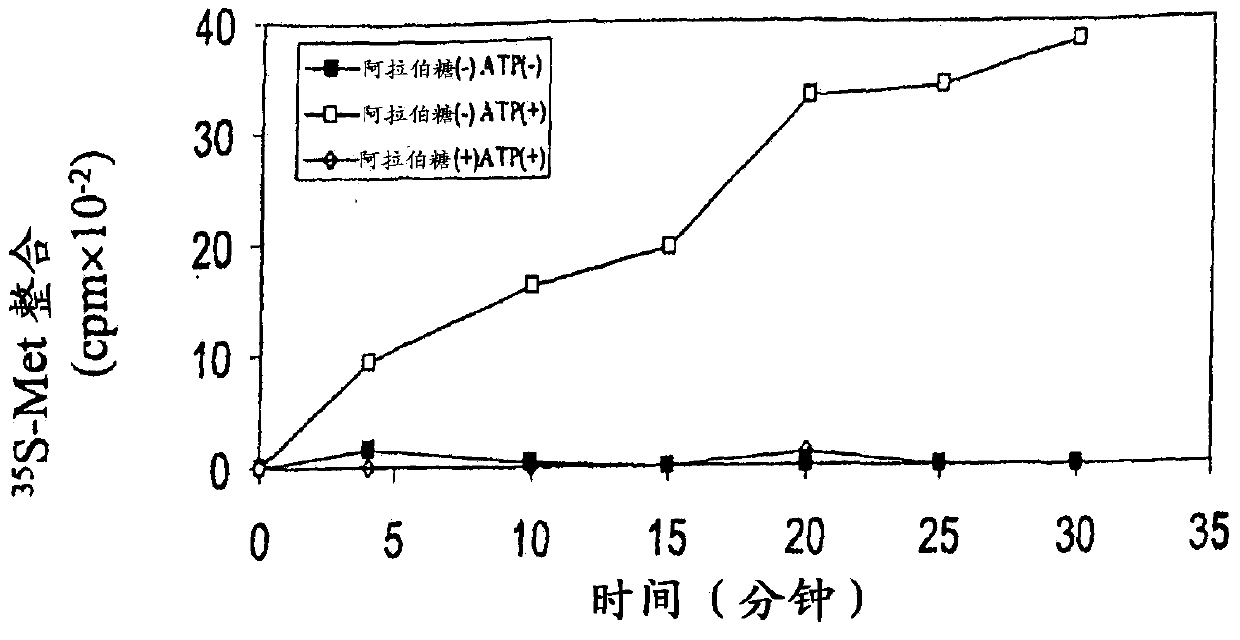
[0285] As described here, treatment with toluene permeabilized E. coli cells, and these cells were used to prove that MazF inhibited translation, but did not inhibit RNA synthesis or DNA replication. Moreover, it was shown that MazF specifically cleaves mRNA between the A and C residues of the ACA sequence in a ribosome-independent manner. Therefore, the present invention proves that MazF interferes with the function of mRNA by cutting the mRNA at a specific site. Therefore, the inventors of the present invention discovered that MazF is a new type of endoribonuclease, and named it "mRNA interfering enzyme" here.
[0286] Materials and Method
[0287] Strains and plasmids. E. coli BL21 (DE3), BW25113 (Datsenko and Wanner, Proc Natl Acad Sci US A97, 6640-5 (2000)) and MRE600 (Swaney et al., Antimicrob Agents Chemother 42, 3251-5 (1998)) were used. The plasmid pET-21cc-MazEF was constructed from the plasmid pET-21cc (Novagen), which was modified to express MazE and MazF (His) under...
Embodiment II
[0326] Importantly, before the discovery of the present invention, the cellular target of MazF has not been identified. As shown here, MazF acts as a highly sequence-specific endoribonuclease, which cuts cellular mRNA at the ACA site. This activity may achieve partial or complete inhibition of protein synthesis in cells. Standard calculations are performed based on the same principle that any one of the four nucleotides is incorporated into each of the three nucleotide positions in the ACA sequence. Based on this standard calculation, the ACA sequence appears in The predicted frequency in RNA transcripts is 1 / 64. It should be understood that some RNA transcripts contain lower or higher frequencies of ACA sequences compared to predicted frequencies. Therefore, the sensitivity of a particular RNA transcript or a family of related RNA transcripts to cleavage by MazF endoribonuclease depends on the frequency of the ACA sequence or MazF target sequence in the transcript. Moreover...
Embodiment III
[0328] As described above, in E. coli, programmed cell death is considered to be mediated through the "addiction component" system, each of which consists of a pair of co-expressed genes, which encode a stable toxin And unstable antitoxin. Their expression is automatically regulated by the toxin / antitoxin complex or only by the antitoxin. When co-expression is inhibited, the antitoxin is rapidly degraded by proteases, allowing the toxin to act on its target. In E. coli, extrachromosomal elements are the main genetic system of bacterial programmed cell death. The most studied extrachromosomal addictive component is the phd-doc on phage P1 (Lehnherr et al. (1993) J Mol Biol 233, 414-428; Lehnherr and Yarmolinsky (1995) Proc Natl Acad Sci USA 92, 3274-3277; Magnuson and Yarmolinsky (1998) J Bacteriol180, 6342-6351; Gazit and Sauer (1999) J Biol Chem274, 16813-16818; Gazit and Sauer (1999) J Biol Chem274, 2652-2657), ccdA-ccdB on the F factor (Tam and Kline (1989) J Bacteriol171...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com