Internal Reference Genes, Primers and Applications of Fluorescence Quantitative Quantification of Oyster oyster poly(i:c) Stress Experiment
A technology of real-time fluorescence quantification and internal reference genes, applied in the field of molecular biology, can solve problems such as differences in the expression of housekeeping genes, and achieve the effect of true and reliable data, high accuracy, and universal application
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
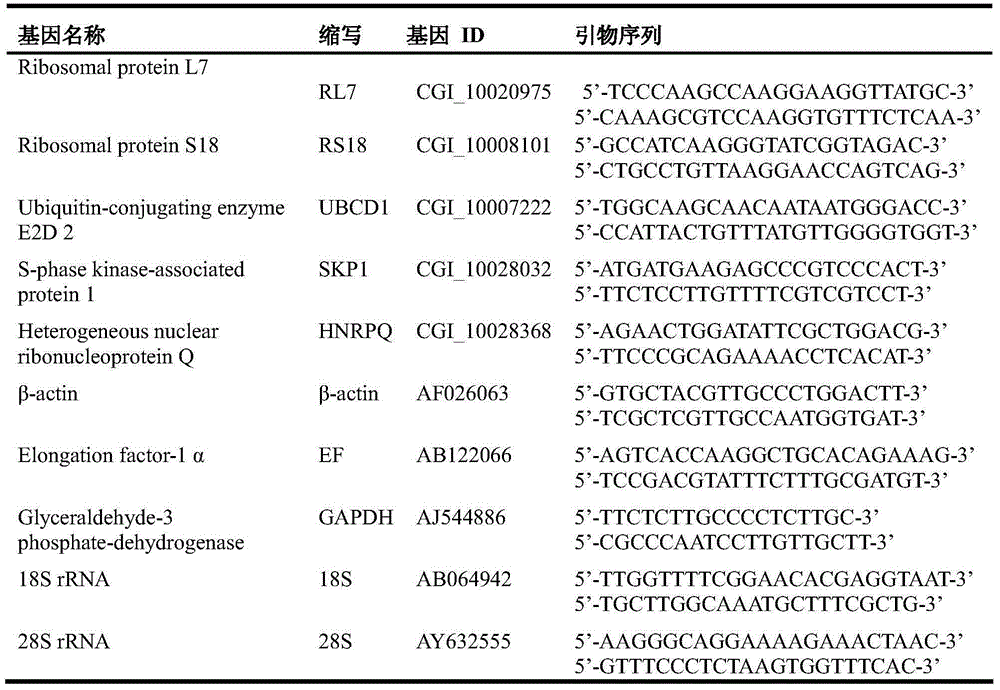
[0064] Selection of candidate internal reference genes: We selected five internal reference genes commonly used in long oyster: Elongationfactor-1α, β-actin, 18SrRNA, 28SrRNA and GAPDH, as well as internal reference genes RS18 and RL7 with stable expression during the development of oyster larvae, and another Three homologous genes of human housekeeping genes SKP1, HNRP and UBCD1 obtained from the oyster genome were used as candidate genes for internal reference genes. The selected internal reference gene sequence (see Table 1)
[0065] Experimental materials used in quantitative experiments: adult healthy long oysters (average shell height 90mm) were purchased from Jiaonan, Qingdao. Before the experiment, they were kept in aerated filtered seawater, and the water temperature was maintained at 18°C±1°C. Poly(I:C) stress experiment: 120 long oysters were equally divided into two groups, 60 in each group. In the experimental group, 60 mice were each injected with 100 μl Poly(I...
Embodiment 2
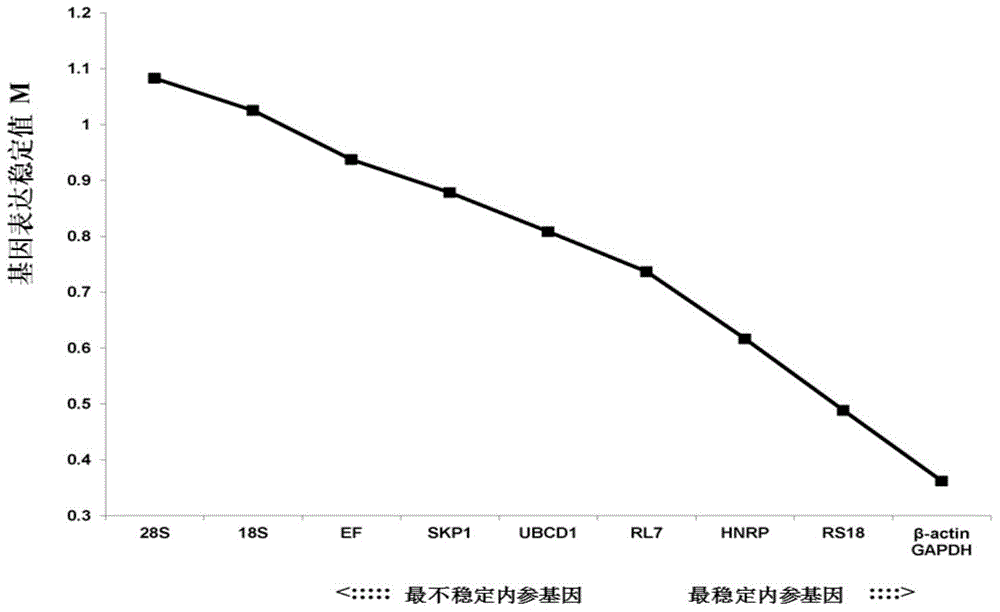
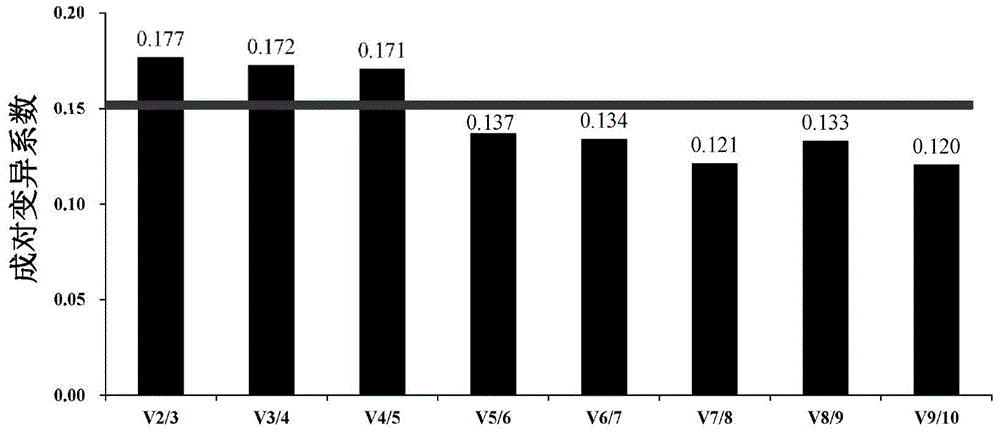
[0068] Evaluate the expression stability of candidate internal reference genes by fluorescent quantitative PCR: design a specific primer pair (see Table 1) according to the candidate gene sequence, perform fluorescent quantitative PCR in Poly(I:C) stress samples, and obtain the expression stability of each candidate gene. The expression data at each stress time point is the Ct value (threshold cycle, the number of cycles experienced when the fluorescent signal in each reaction tube reaches the set threshold value). According to the Ct value, the stable value of each candidate gene was calculated with the GeNorm software commonly used for internal reference screening. The smaller the stable value, the more stable the expression of the gene. In addition, GeNorm software through the pairwise variation coefficient V n / V n+1The analysis can determine the optimal number of housekeeping genes. Vandesompele et al. suggested setting 0.15 as the threshold of the pairwise coefficient ...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com