Pavlova-viridis-derived delta-6 desaturase new gene
A desaturase and gene technology, applied in the field of desaturase genes and their separation, can solve the problems of different enzyme activities and functions, differences in sequence structure, etc.
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment Construction
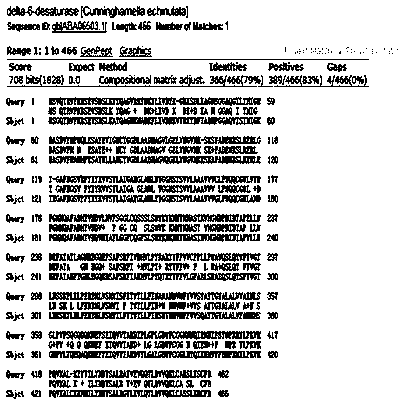
[0019] The isolation process of Pavlova viridis delta-6 desaturase gene is as follows:
[0020] (1) Obtain the total mRNA of Pavlova viridis using conventional mRNA extraction and separation means in the art, and use primer OligodT to reverse transcribe it into cDNA by conventional means in the art;
[0021] (2) Using cDNA as a template, design primers P6F and P6R according to the conservatism of the delta6 desaturase homologous gene to amplify the gene in the conserved region. The primer sequence is: P6F: 5'-GTTCCTCGCTGTCGTGGTGGA-3'; P3R: P3R: 5'-TGTTGTGACCGCCGCAGAACCA-3'; PCR reaction conditions: 94oC 3 min; 94oC 30 s, 60oC 30 s, 72oC 1 min, 30 cycles; 72oC 10 min;
[0022] (3) The amplification reaction at the 3' end was carried out using cDNA as a template and using the SMART RACE cDNA amplification kit. The amplification primers used were: UPM (10×universal primer mix); des6-3 TTGGATCTCGTTCATCACTTG. The gradient PCR reaction program is: 94oC 3 min; 94oC 30 s, 72oC 3 min,...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com