Detection device and method for gene fusion
A detection method and gene fusion technology, applied in the field of bioinformatics, can solve problems such as verification, high false positives, and inability to determine breakpoint information
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0076] Example 1 according to figure 2 In the process shown, the whole genome sequencing data of simulated rice is compared with the rice reference genome based on the paired-end sequence using the BWA sequence alignment software, as shown in Figure 6a As shown, the comparison result in BAM format is obtained. For an example, see Figure 6b .
[0077] in, Figure 6a The coor in represents the ruler; ref represents the reference sequence; r001 to r004 represent the sequences to be compared; Figure 6b Indicates the results after r001 to r004 are aligned to the reference sequence, the first line and the second line are the titles of the BAM file. The remaining columns respectively represent: 1: sequence number; 2: FLAG value of sequence alignment, expressed in binary mode, reflecting various information of sequence alignment to reference sequence, for example, it can reflect whether the sequence alignment is Success, forward and reverse strand information of the alignment,...
Embodiment 2
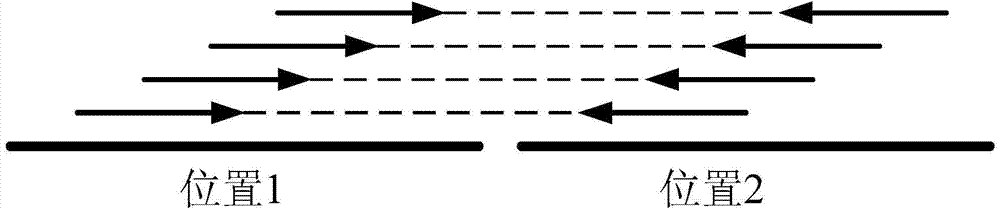
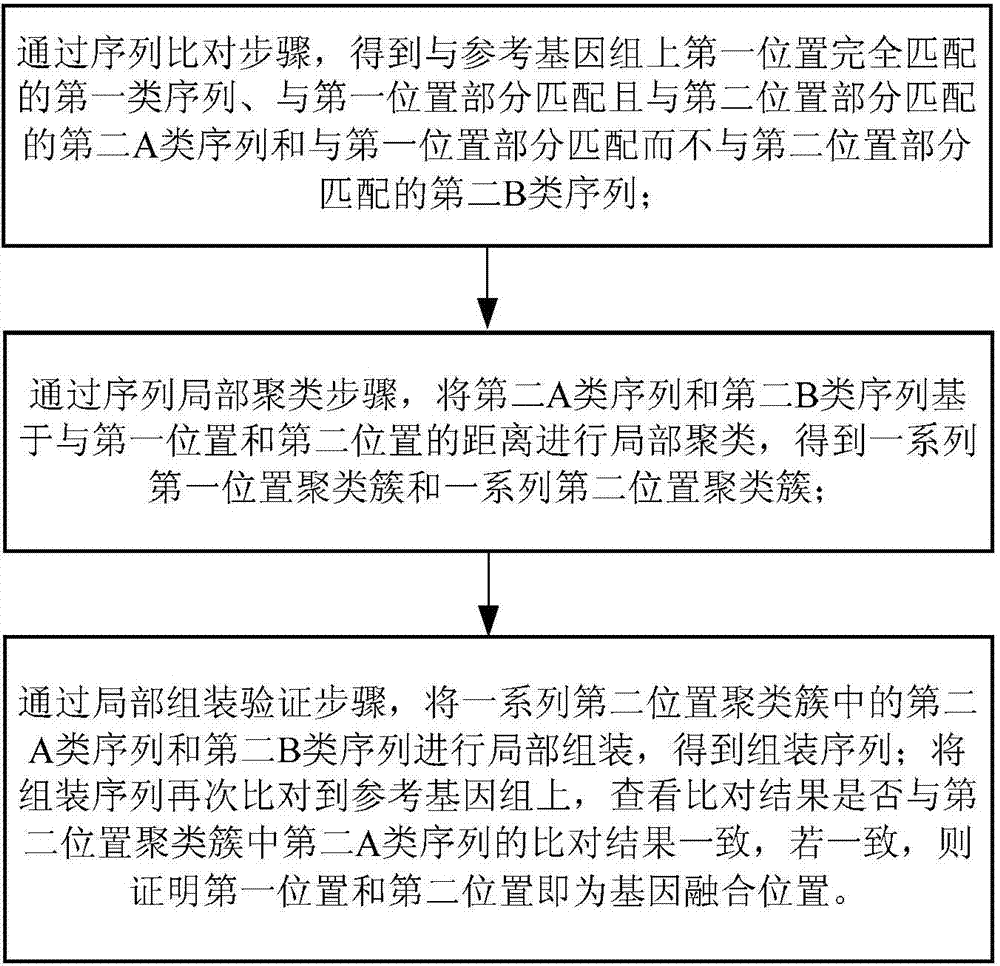
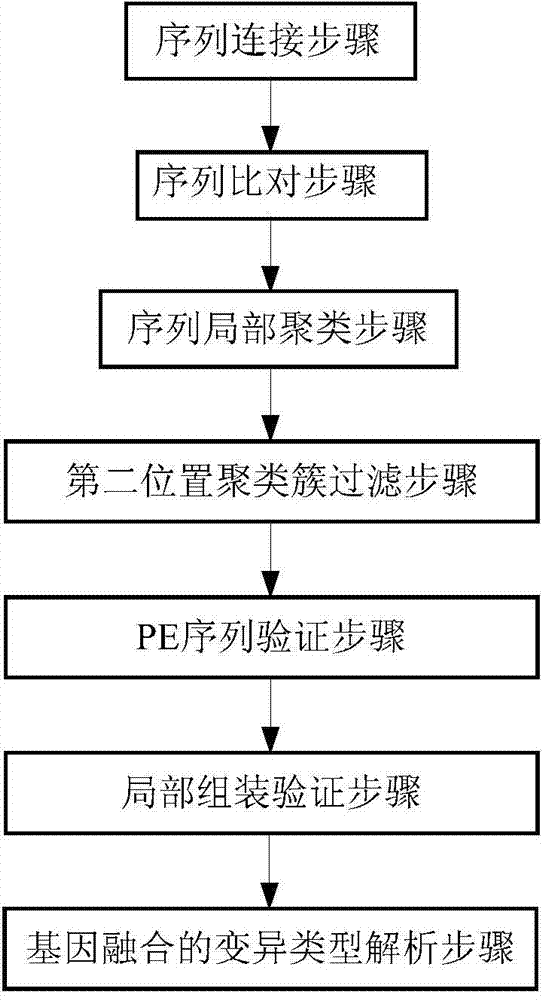
[0082] Example 2 according to image 3 In the shown process, firstly, the fragments with multiple different overlapping sequences in the simulated sequence are connected to obtain multiple longer connected fragments; the above-mentioned connected fragments and the simulated sequencing sequences without overlapping Compare the reference genome sequence to obtain the comparison results of the connected fragments and the non-connected fragments; combine the comparison results of the connected fragments and the non-connected fragments to obtain the total comparison result; sort the total comparison results , to obtain the first type of fragments that completely match the first position of the reference genome sequence of rice and the second type of fragments that do not completely match the first position; the second type of fragments include the second A type of fragments that can be aligned to the second position and a second class B fragment that is not aligned to a second posi...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com