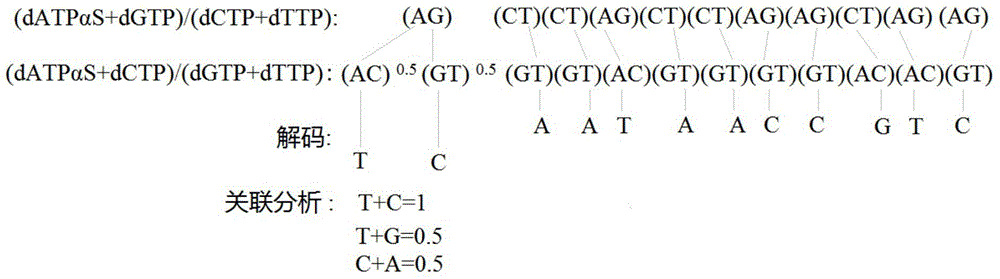Two-nucleotide synthetic sequencing analysis method for multi-template PCR product
A two-nucleotide, sequencing-by-synthesis technology, used in biochemical equipment and methods, and microbial determination/inspection. Effect
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
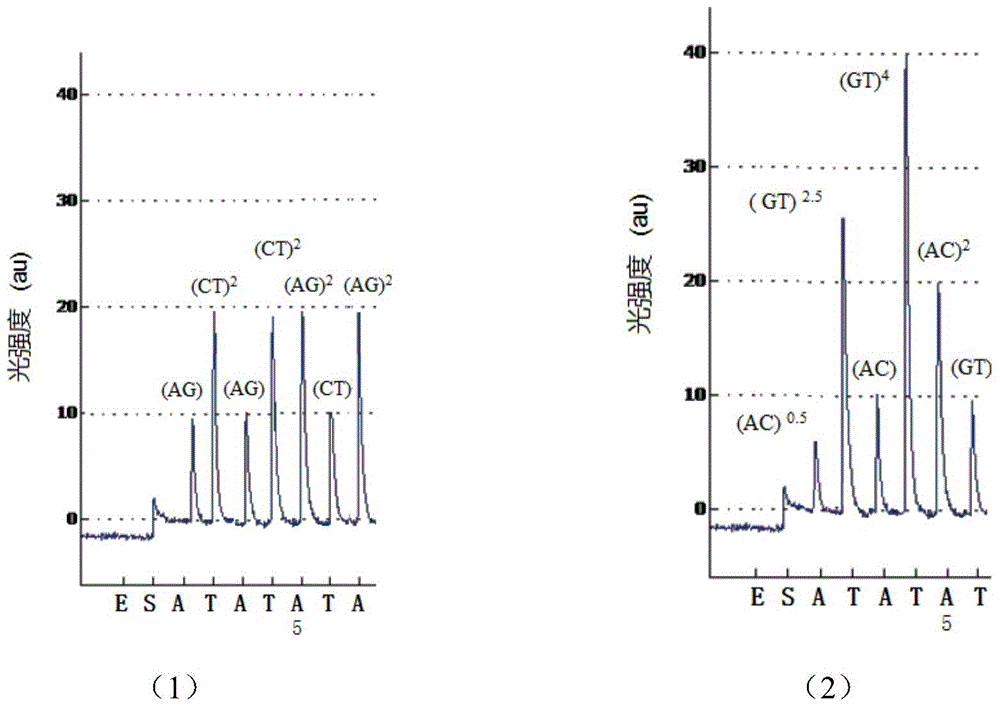
[0047] Embodiment 1: The human sample comprises the PCR product DNA template analysis method of MMP7 (U25346) gene comprising a SNP (C / T, rs11568819 site, and specific method comprises:
[0048] (1) Genomic DNA in peripheral blood was extracted from the blood sample using traditional protein kinase K and phenol / chloroform extraction methods;
[0049] (2) Combine PCR primers 5'- agtctacaga actttgaaag tatgtg -3', 5'-biotin- ctatgagagc agtcatttga ctttg -3' with 200 mg genomic DNA, 0.2 mM dNTP, 1 U Taq DNA polymerase, 1× amplification buffer , 1.8 mM MgCl 2 The 50 μL PCR amplification system was used for amplification. The amplification conditions were: initial denaturation at 94 °C for 5 min; 35 thermal cycles: denaturation at 94 °C for 30 s, annealing at 61 °C for 45 s, and extension at 72 °C for 45 s; Extend at 72 °C for 7 min;
[0050] (3) Divide the PCR amplification product evenly into two parts, and carry out the operations of steps (4) to (6) respectively;
[0051] (4) ...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com