DNA sequence similarity analysis method based on S-PCNN and Huffman encoding
A DNA sequence and Huffman coding technology, applied in the field of bioinformatics, can solve problems such as accumulation errors, low DNA sequence discrimination, similarity analysis errors, etc.
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment Construction
[0020] The present invention will be described in further detail below in conjunction with the accompanying drawings and embodiments.
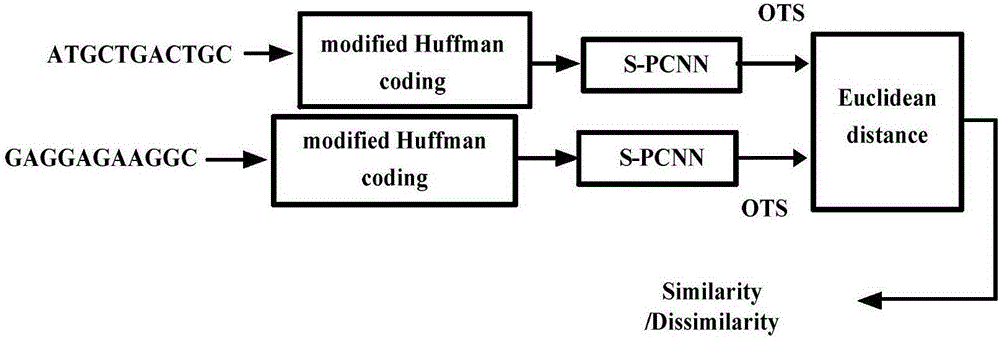
[0021] The basic idea of the present invention is: count the number of 64 kinds of triplet codons in the DNA fragment, and obtain its occurrence probability, then carry out Huffman coding to the probability of 64 kinds of triplet codon occurrences, and then convert each code into Decimal numbers, and normalize the encoding to the range of 0 to 1, replace the triplet codon character sequence of DNA with a digital sequence, and then send the encoded DNA digital sequence to the S-PCNN model for clustering calculation to obtain DNA An oscillatory time series (OTS) of a sequence. Finally, the Euclidean distance between the oscillation time series of different DNA sequences is calculated, and the degree of kinship between species is judged by the Euclidean distance. Its method flow chart is as follows figure 1 shown;
[0022] Specifically, the ...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com