Cardiotonic steroid antagonists and related methods
A cardiovascular, non-natural technology, applied in chemical instruments and methods, biochemical equipment and methods, pharmaceutical formulations, etc., to solve problems such as specificity loss
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0077] Example 1 - The second cytoplasmic domain of Na / K-ATPase as a modulator of the Src SH2 domain.
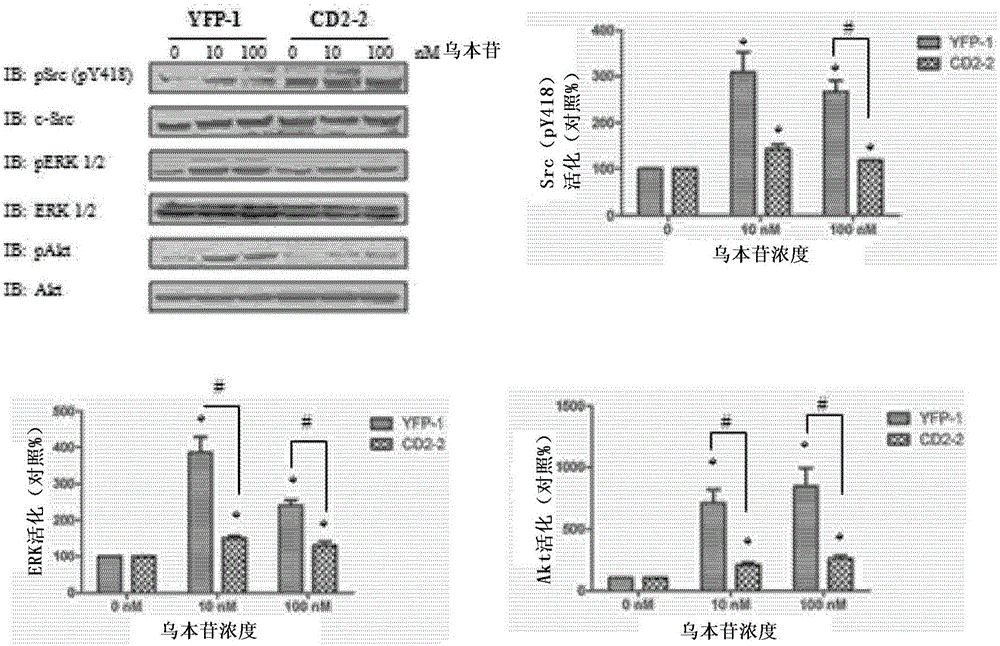
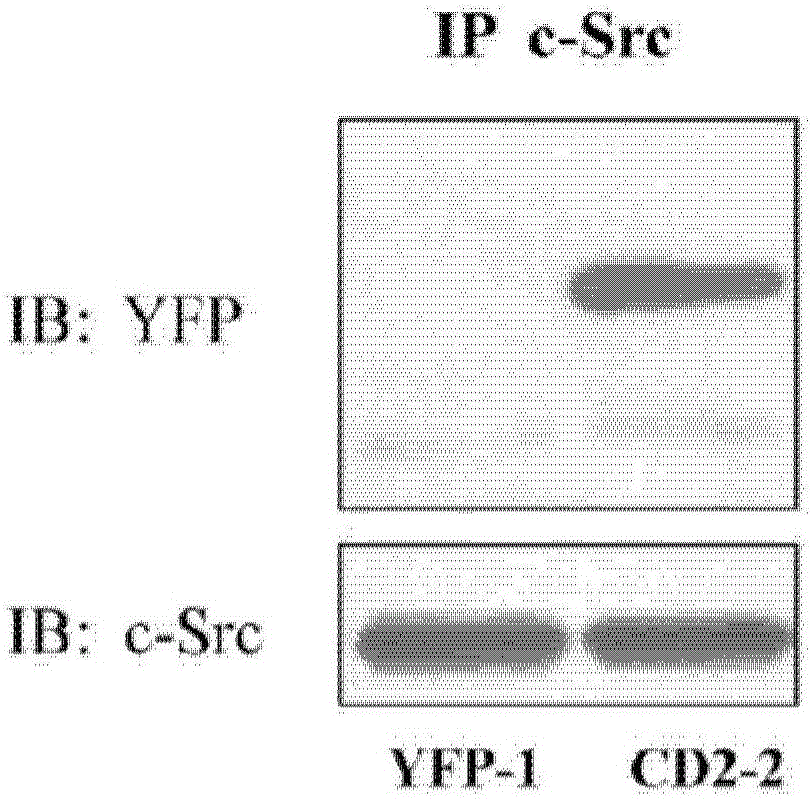
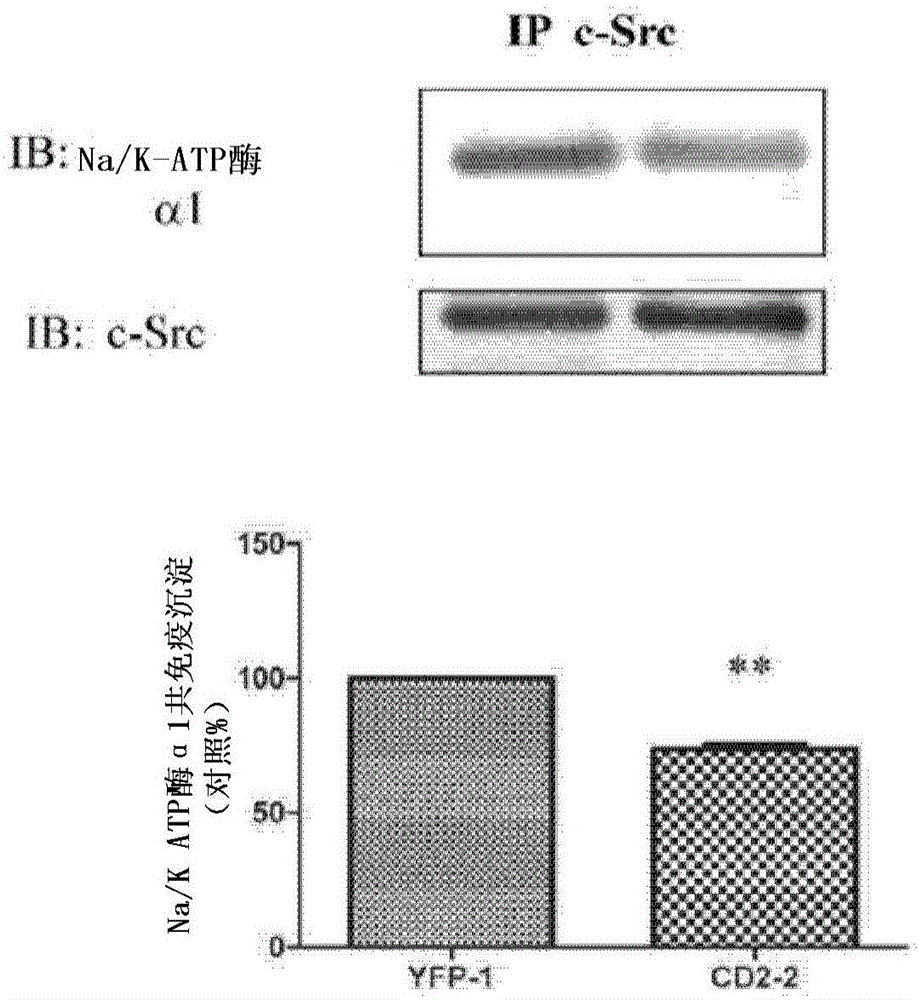
[0078] Na / K-ATPase directly interacts with Src kinases through two domain-domain interactions (Tian, Cai et al. 2006; Ye, Li et al. 2011). Specifically, the second cytoplasmic domain (CD2) of Na / K-ATPase binds the Src SH2 domain, and the N domain of Na / K-ATPase binds the Src kinase domain. Conjugation of cardiotonic steroids such as ouabain disrupts N domain / kinase domain interactions, whereas CD2 / SH2 interactions are usually constitutive. In this regard, because CD2 binds the Src SH2 domain, and the SH2 domain functions in targeting Src kinases to specific signaling complexes, it is believed that ectopically expressed CD2 will act as the Na / K-ATPase / Dominant-negative mutant of the Src receptor complex and thereby inhibits ouabain-mediated cell signaling. To test this notion, CD2 was expressed in LLC-PK1 cells as a yellow fluorescent protein (YFP) fusion protein, and ...
Embodiment 2
[0081] Example 2 - Development of Peptide Antagonists for Disruption of CD2 / SH2 Interaction.
[0082] In light of the foregoing findings, experiments were then performed to develop highly specific peptide antagonists for disrupting the CD2 / SH2 interaction to inhibit cardiac steroid-mediated signaling in cells. It has been observed that the Src SH2 domain displays preferential binding to phosphorylated tyrosine residues. like Figure 4A CD2 from the α1 subunit of Na / K-ATPase (mammals; SEQ ID NO: ) contains only a single tyrosine at position 260 (in unmodified Na / K-ATPase), as shown in To determine whether this tyrosine can be phosphorylated in CD2 expressing cells. like Figure 4B As depicted in , CD2 can be phosphorylated when it is expressed in cells. Figure 4C It was further shown that in normal LLC-PK1 cells, tyrosine 260 of the full-length Na / K-ATPase was phosphorylated. Furthermore, this phosphorylation can be stimulated by ouabain, since ouabain increases phospho...
Embodiment 3
[0083] Example 3 - Affinity of Peptide Antagonists
[0084]A peptide derived from CD2 called CD2C2 (see, e.g., SEQ ID NO: 1 in PCT Application No. PCT / US2011 / 021130, incorporated herein by reference) was previously shown to be effective when assayed using a reconstituted in vitro system Glycoside antagonists. In view of the above new findings and previous information on CD2C2, experiments were then performed to assess whether the same peptide (see SEQ ID NO: 1 of the present application) that Y260 is phosphorylated has more Higher affinity. Briefly, to perform these experiments, and to make the peptide permeable to cell membranes, the peptide of SEQ ID NO: 1 was attached to a TAT tag as previously done with other Na / K-ATPase-derived peptides (Li, Cai et al. 2009). The new peptide was designated pNaSH2 (SEQ ID NO:3). For comparison and contrast, CD2C2 with the same TAT tag was also synthesized and designated NaSH2 (SEQ ID NO:4) to indicate that the peptide was not phospho...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com