alpha 1 Three-dimensional crystal structure model of -ar subtype protein and its establishment method
A technique for 1-AR, crystal structure
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
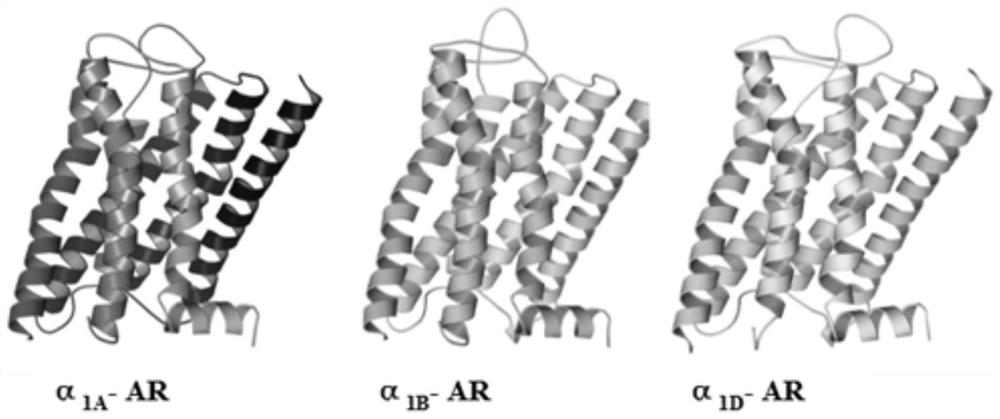
[0032] This embodiment provides a 1 -A three-dimensional crystal structure model of the AR subtype protein and its establishment method, the establishment method comprising the following steps:
[0033] S1: with α 1 -AR isoform protein alpha 1A -AR,α 1B -AR and α 1D The amino acid sequence of -AR was used as a probe, and the protein crystal database was searched by the ProteinBlast program to obtain the homologous template protein β 2 -AR (Code: 2RH1);
[0034] Obtain three subtypes of α1-adrenoceptor (α1-AR) through swiss-prot (http: / / www.uniprot.org / ) 1A -AR, α 1B -AR, α 1D -AR) amino acid sequence, swiss-prot sequence numbers are: P35348, P35368 and P25100 respectively. 2RH1 is a β2 adrenoceptor, and is a member of the GPCR receptor A family with the α1 adrenoceptor to be modeled ; and the β2 adrenergic receptor is an important template for studying GPCRs. It belongs to the same family of adrenoceptors as the α1 adrenoceptor to be modeled. It has higher scores and ...
Embodiment 2
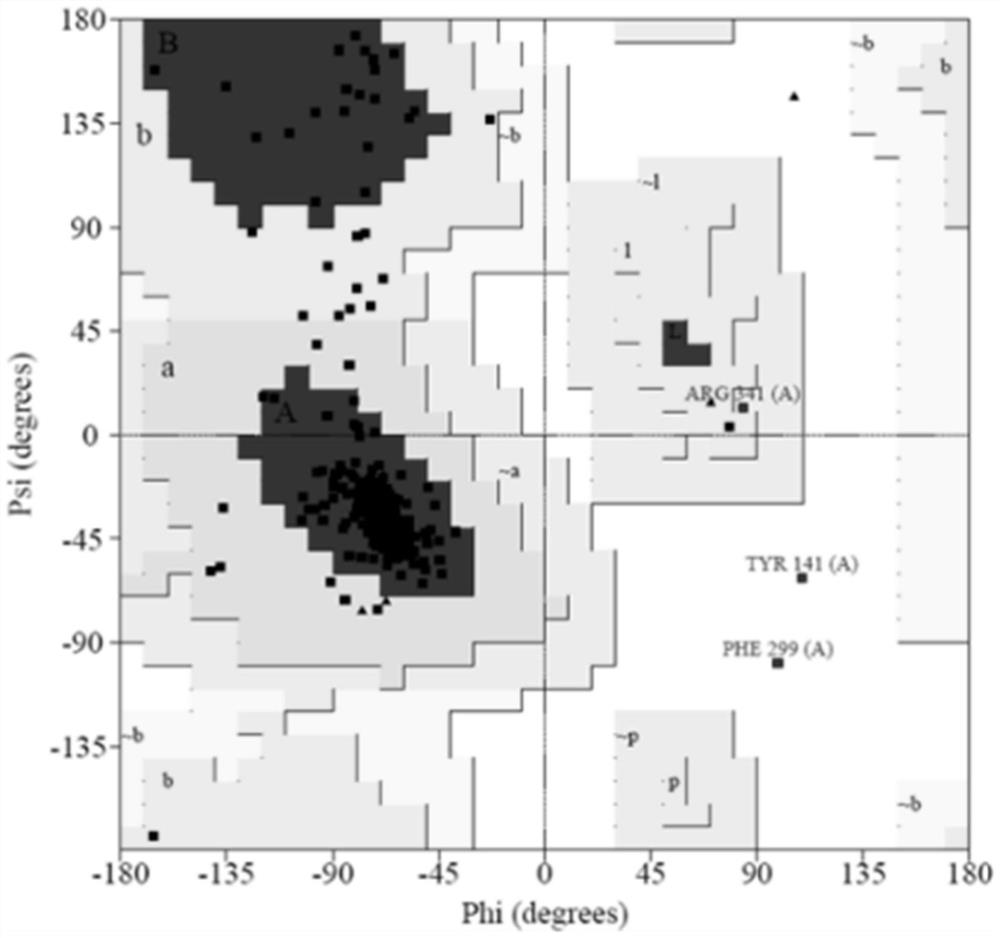
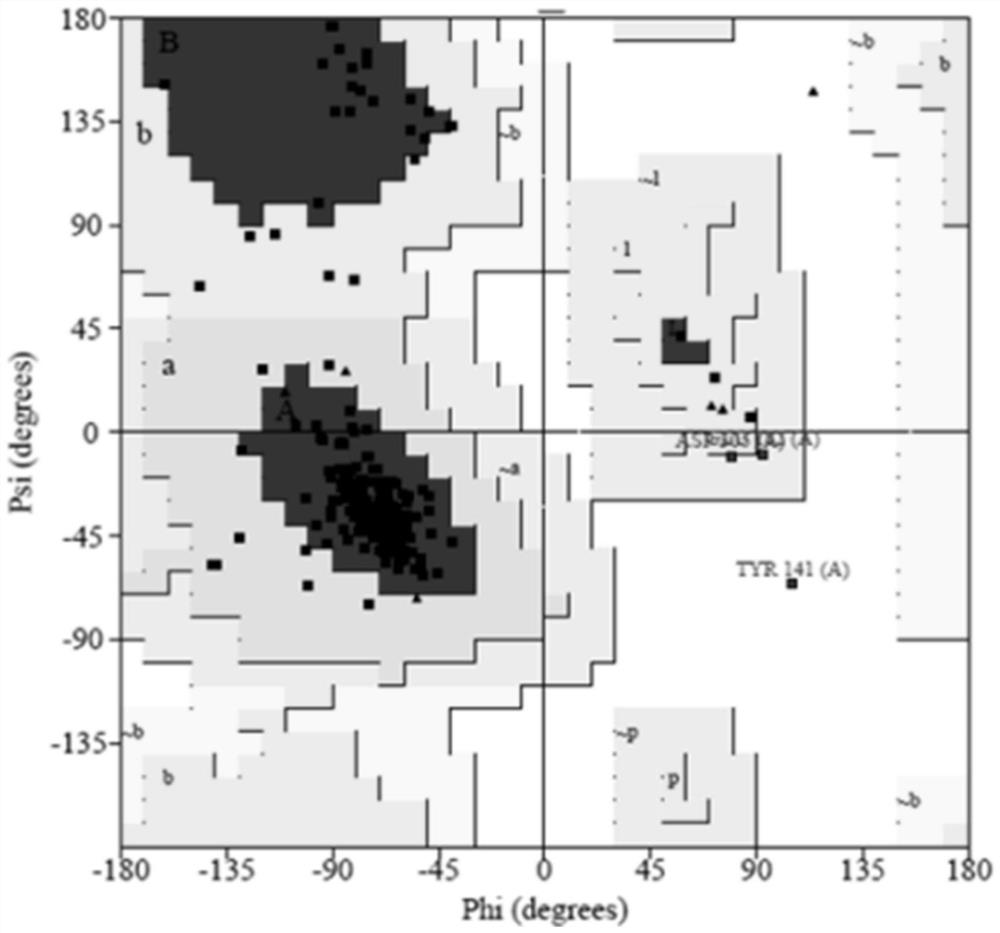
[0043] This embodiment provides a 1 -Evaluation method of three-dimensional crystal structure model of AR subtype protein: using Procheck to obtain α obtained in Example 1 1A -AR,α 1B -AR and α 1D - The physiological and chemical parameters of the three-dimensional crystal structure model of AR are evaluated and analyzed, and the obtained Ramachandran plot can be used to illustrate the rotation degree of the bond between the α-carbon atom and the carbonyl carbon atom in the peptide bond in the protein or peptide stereostructure, and the α-carbon atom The rotation degree of the bond between the nitrogen atom and the nitrogen atom is mainly used to indicate the allowed and disallowed conformations of amino acids in proteins or peptides. More than 90% of the amino acids in the obtained Ramachandran plot are located in the core region, which conforms to the in vitro chemical energy rule.
[0044] The Procheck result of the present invention is as figure 2 , image 3 with F...
Embodiment 3
[0047] This embodiment obtains α according to embodiment one 1A -AR,α 1B -AR and α 1D -A 3D crystal structure model of AR, based on where the ubiquitous binding pocket of the GPCR is located, modeling α 1 -AR three subtypes α 1A -AR, α 1B -AR and α 1D - AR possible binding pockets and find α 1A -AR, α 1B -AR and α 1D - The key residues of the three subtypes of AR, the specific results are as follows Figure 8 , Figure 9 with Figure 10 shown.
[0048] alpha 1A – AR, α 1B -AR and α 1D -The key residues of the three subtypes of AR include: the amino acid residues at position 193 are Ile 193, Val 193 and Ile 193, the amino acid residues at position 293 are Met 293, Leu 293 and Leu 293, and the amino acid residues at position 302 are The amino acids at positions are Lys302, Leu302 and Leu302, respectively. Although alpha 1 -The amino acid residues around the three subtypes of AR are very similar, but each subtype still has its own unique amino acid residues, which...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com