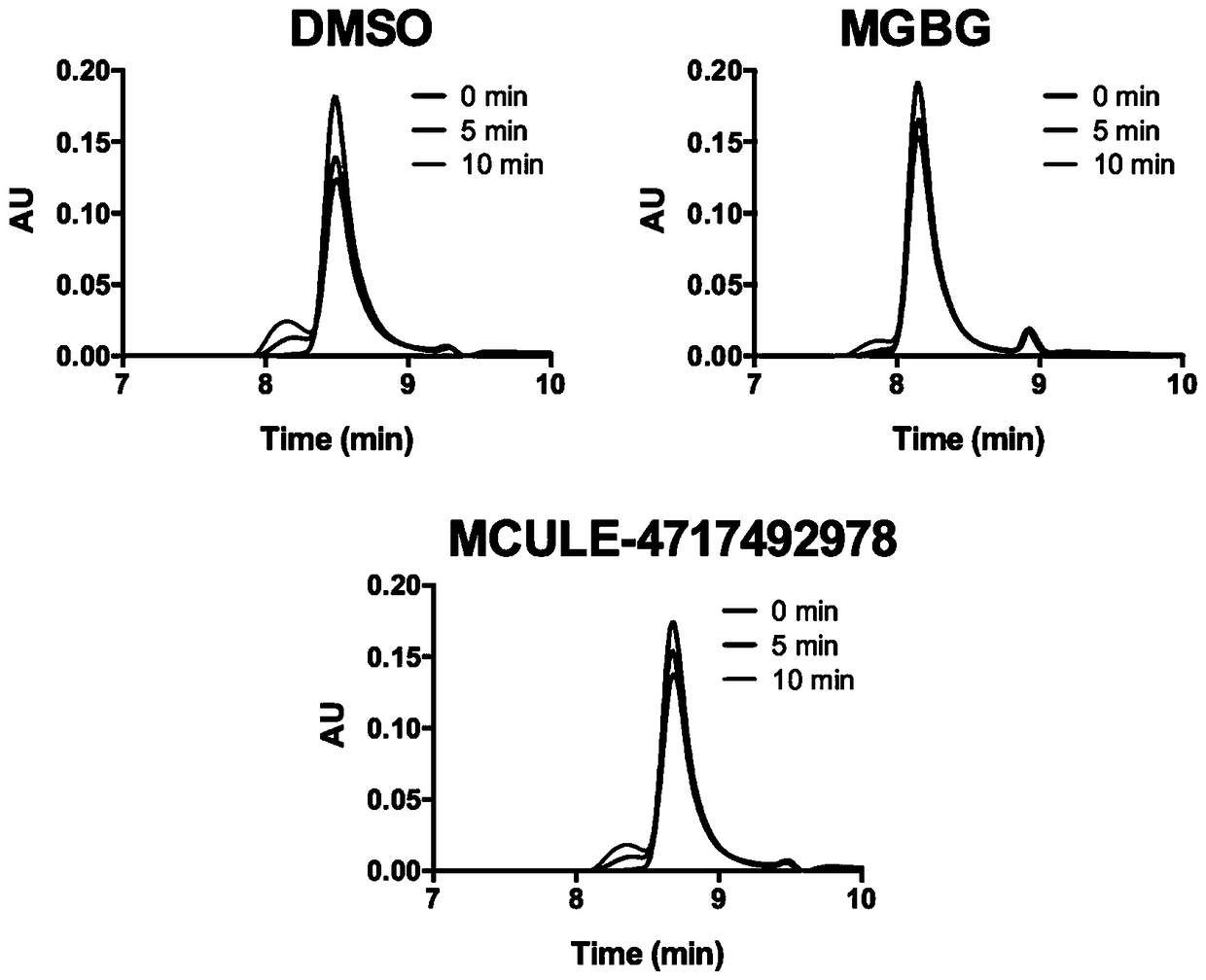
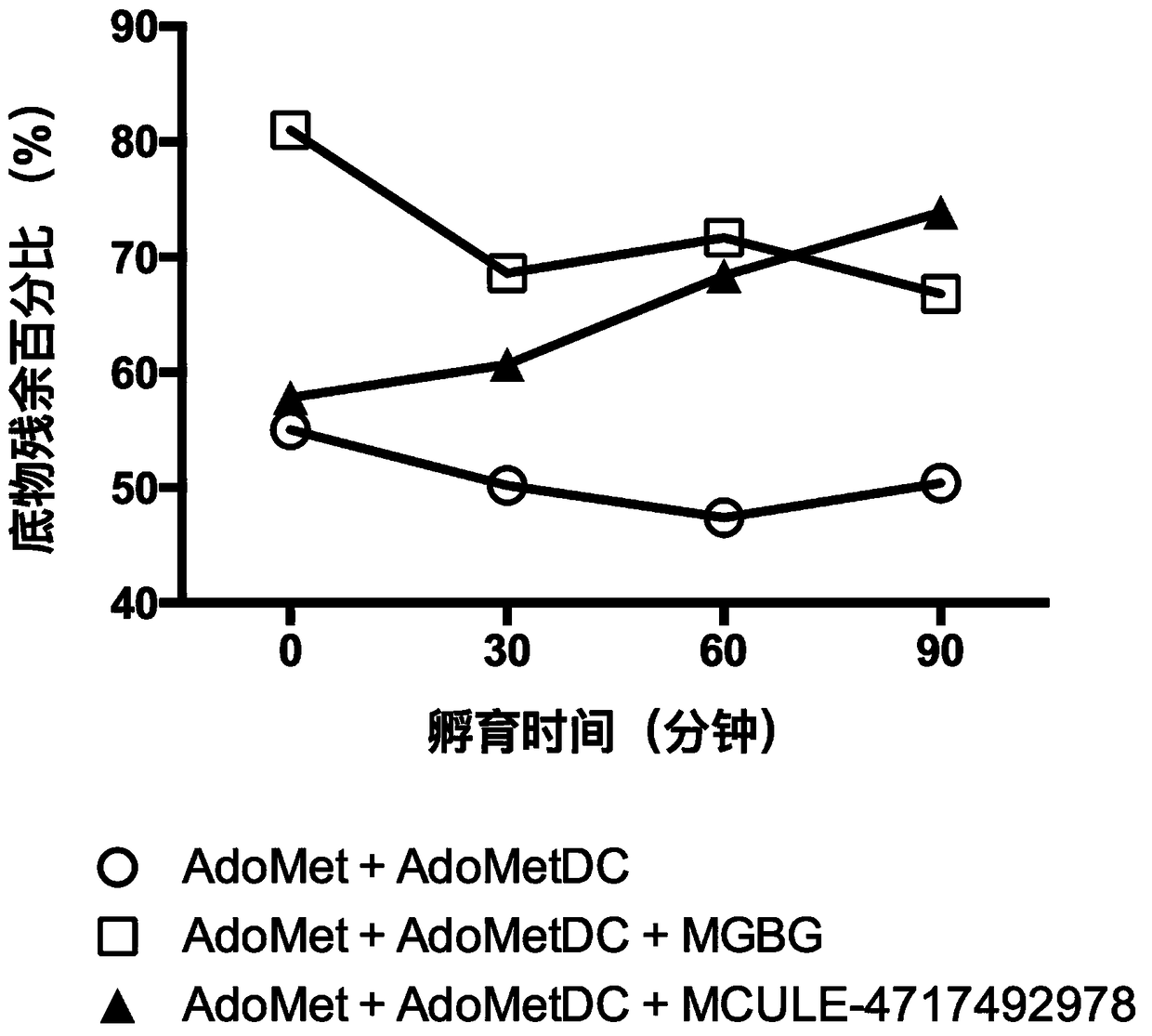
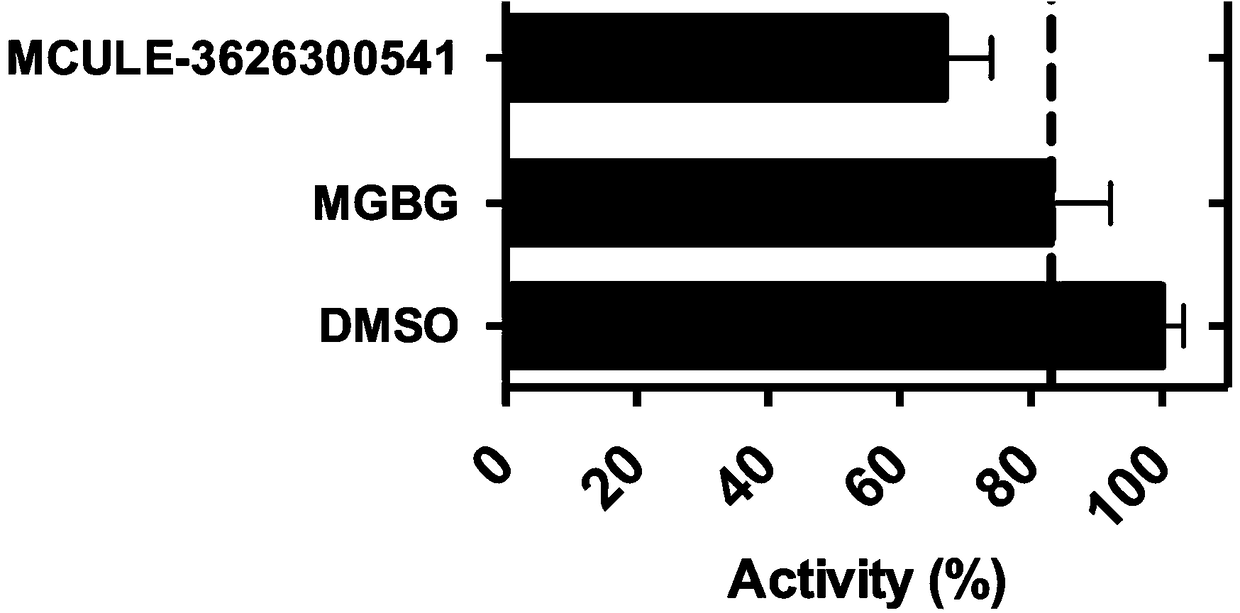
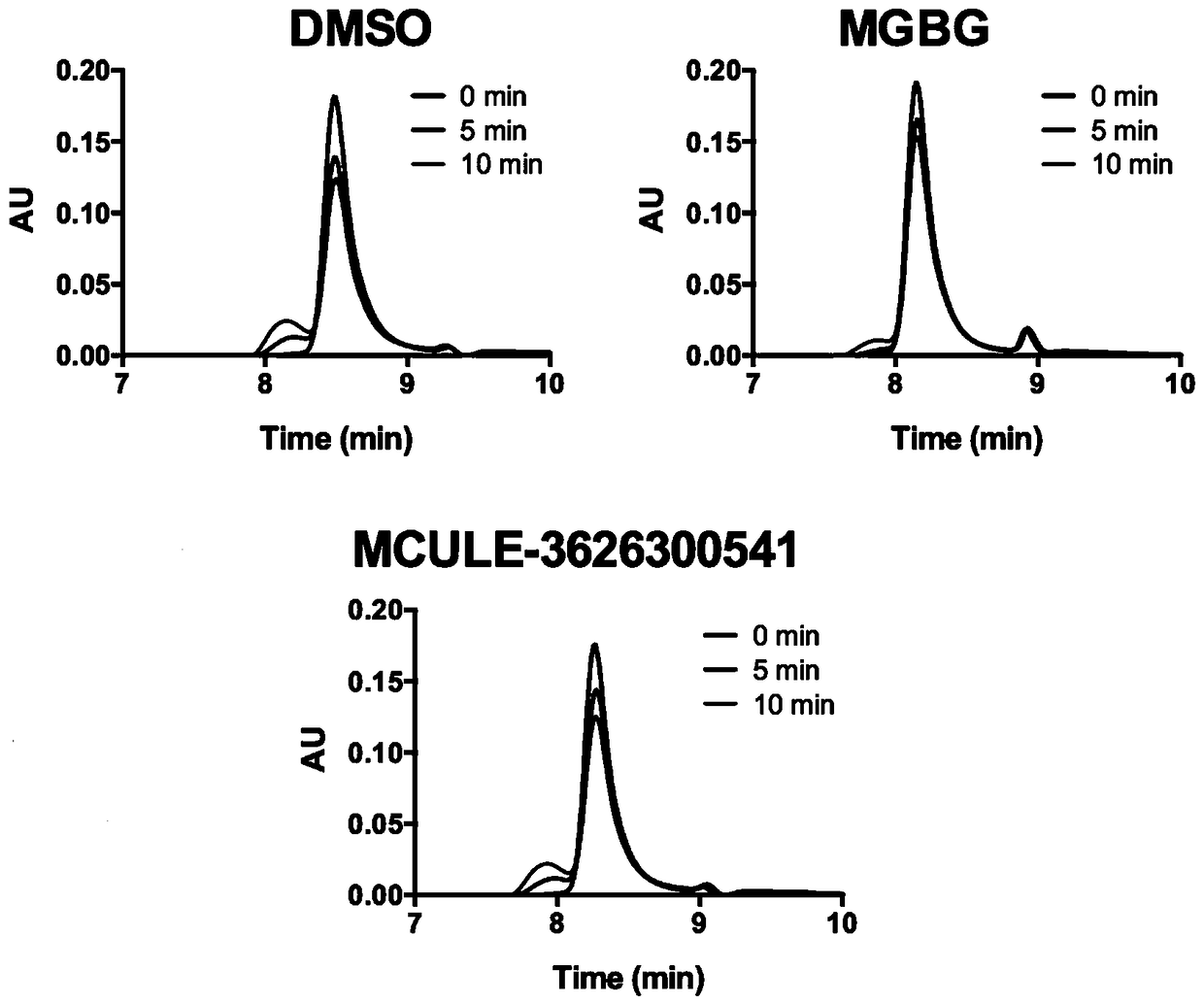
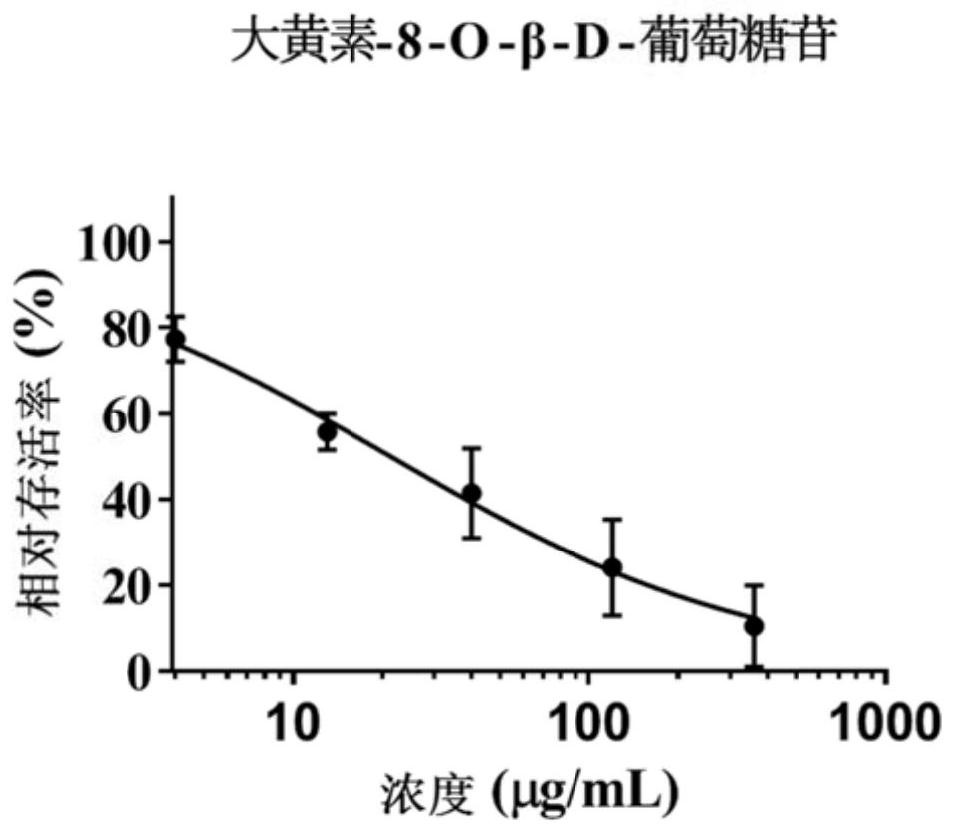
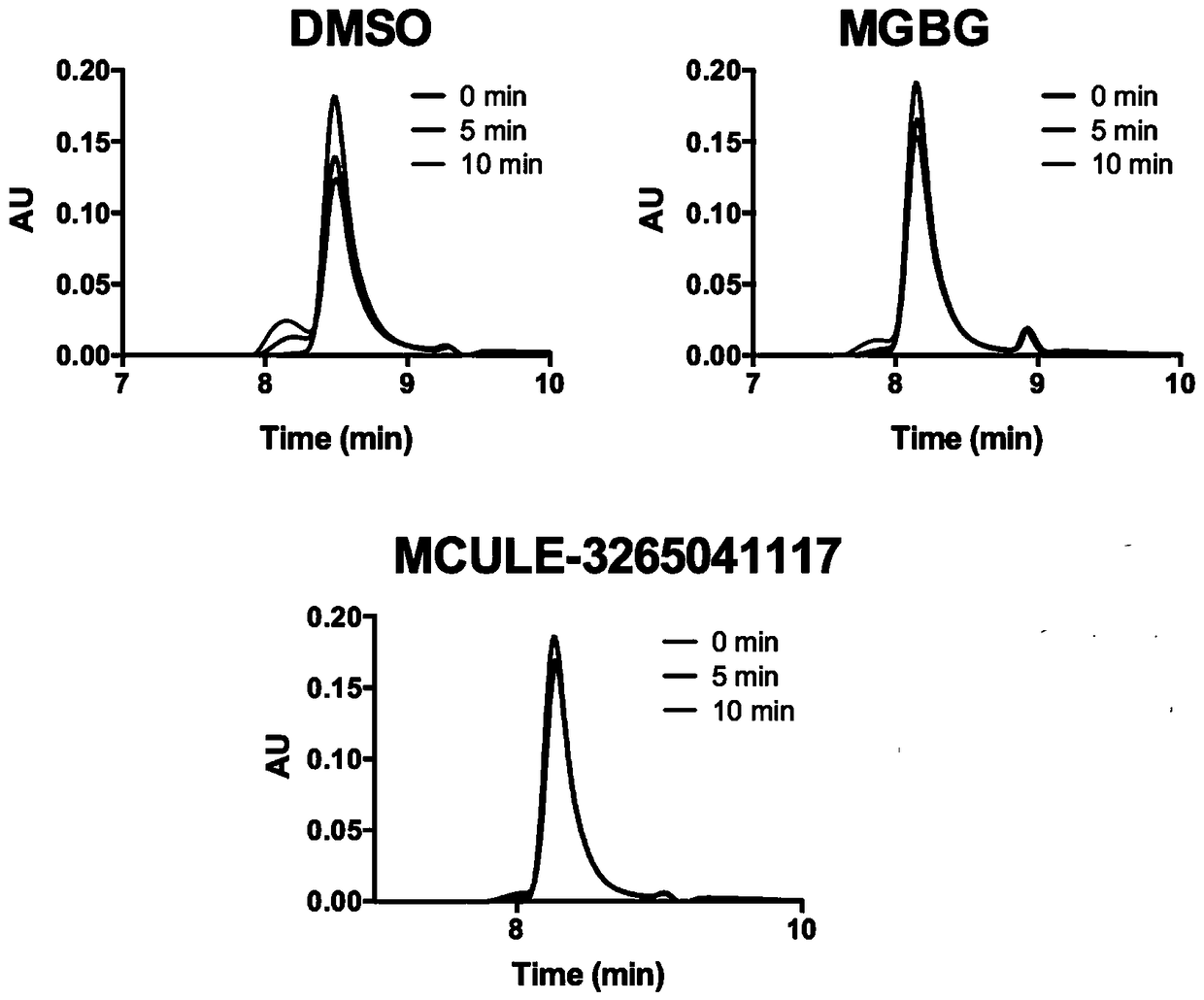
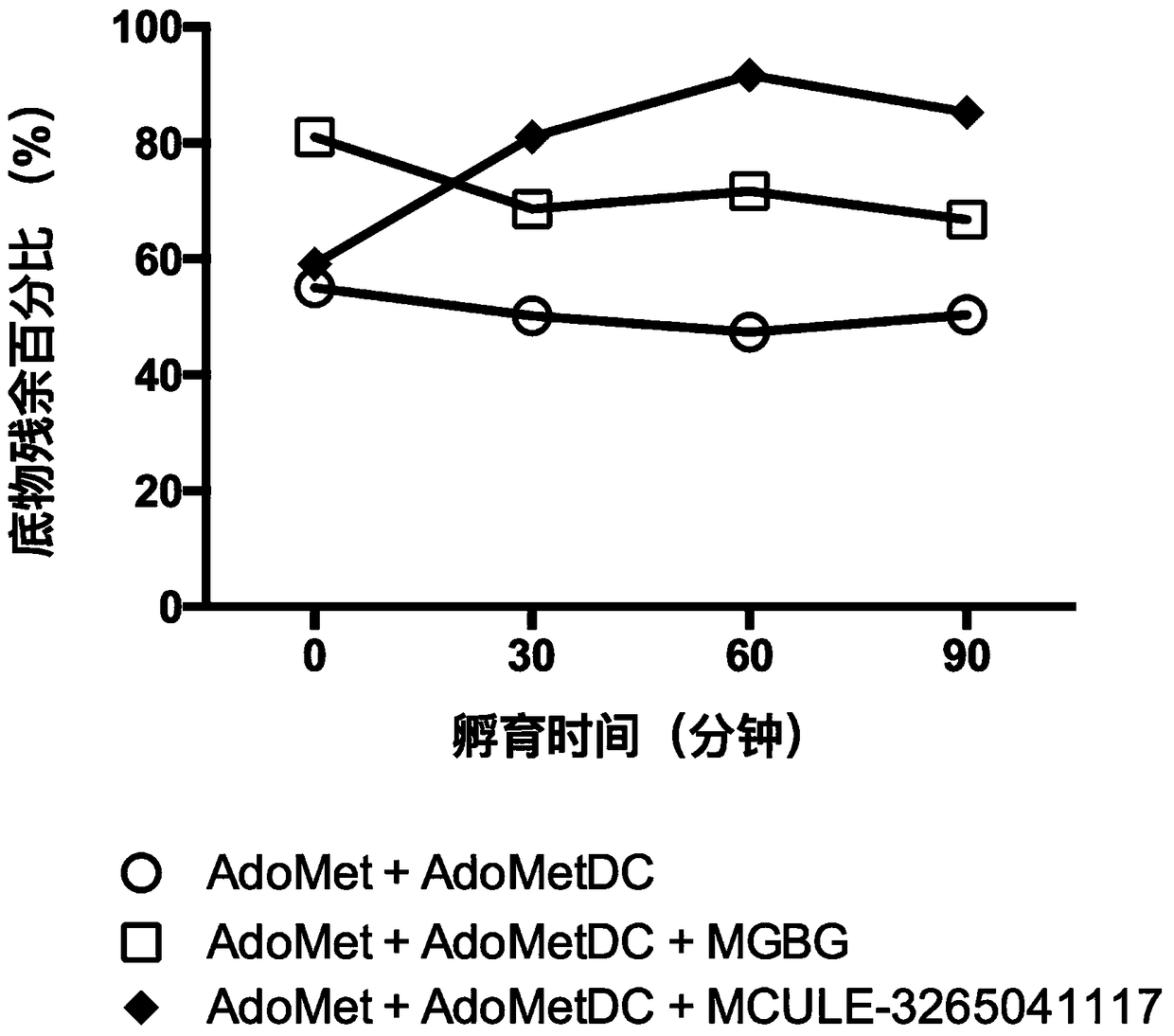
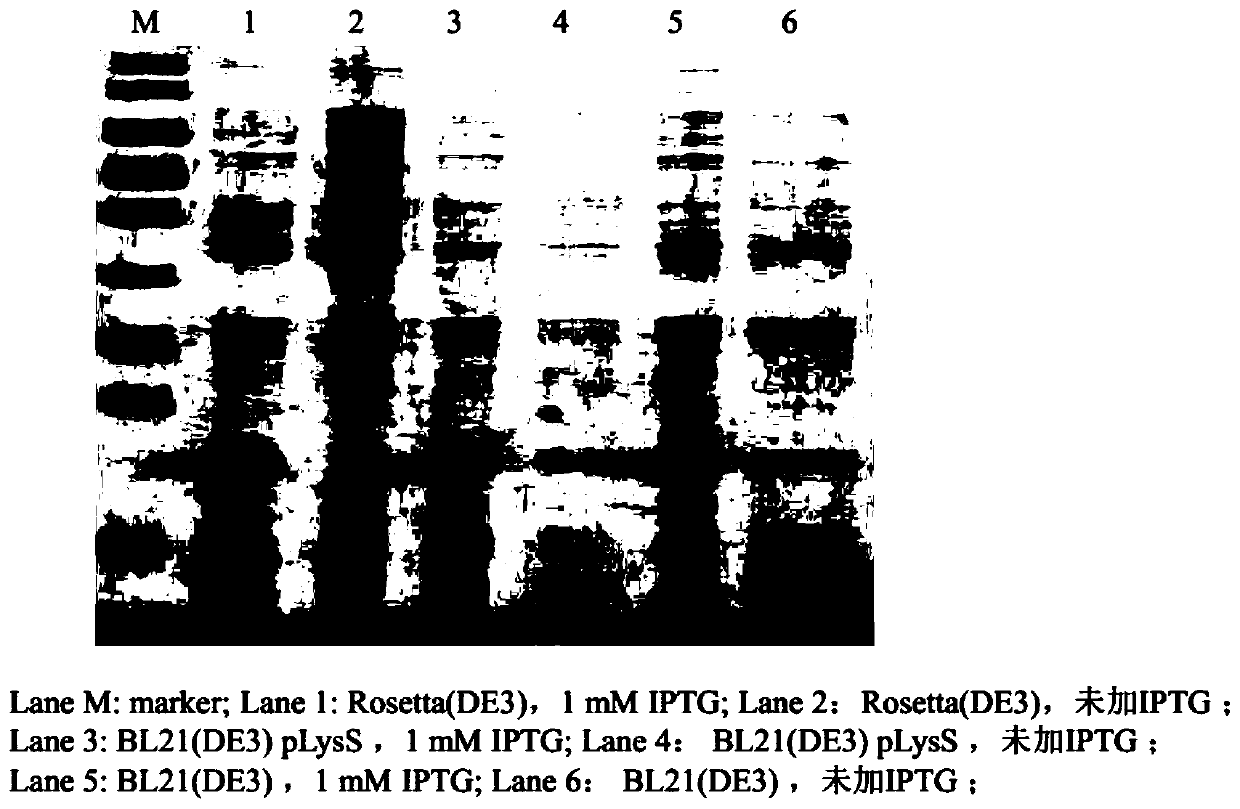
Patents
Literature
32 results about "Protein crystal structure" patented technology
Efficacy Topic
Property
Owner
Technical Advancement
Application Domain
Technology Topic
Technology Field Word
Patent Country/Region
Patent Type
Patent Status
Application Year
Inventor
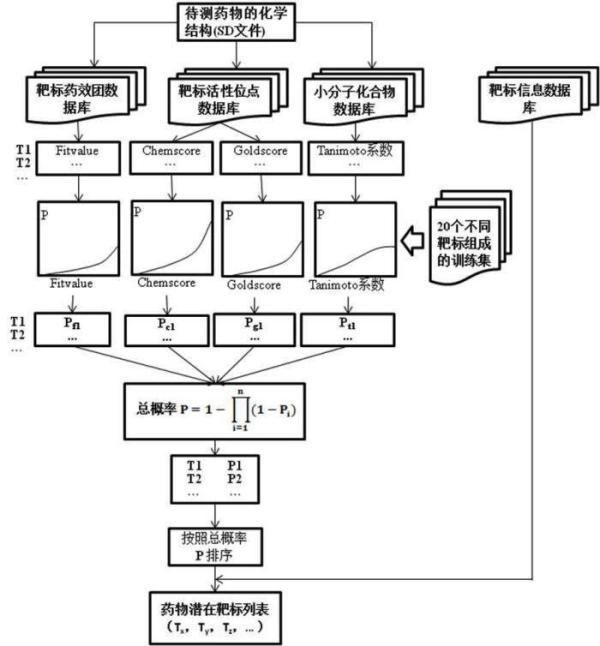
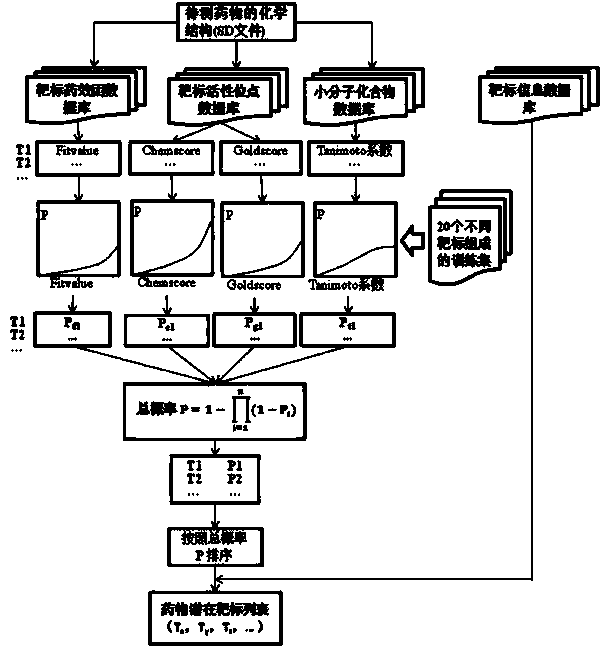
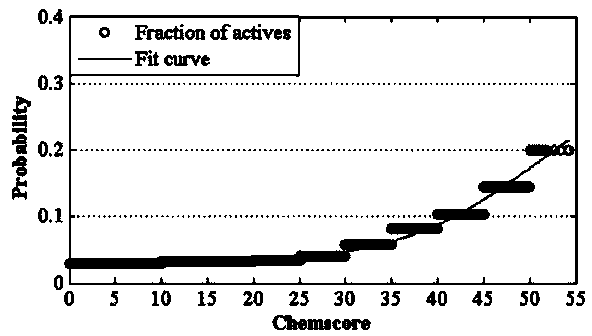
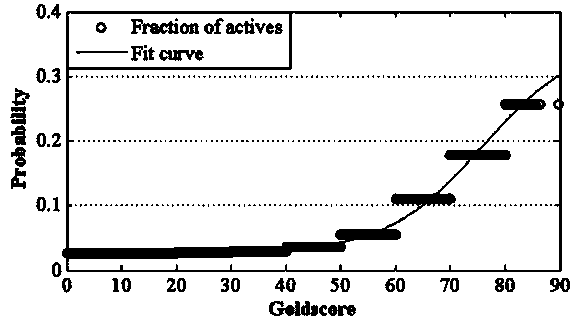
Construction and prediction method of integrated drug target prediction system
InactiveCN102663214AOvercoming the problem of low forecasting accuracySpecial data processing applicationsPharmacophoreEngineering
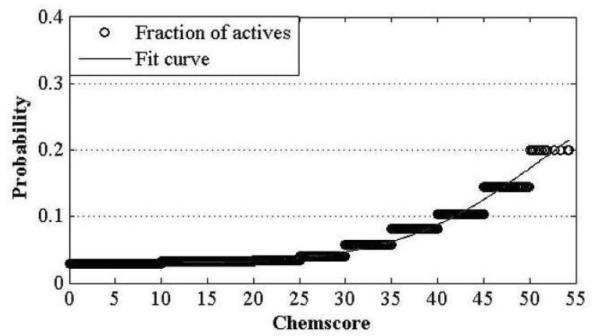
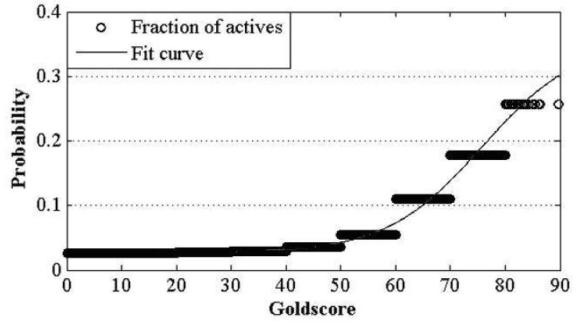
The invention discloses a construction and prediction method of an integrated drug target prediction system. The method comprises the following steps of: analyzing a protein crystal structure database; selecting the protein bound with the drug-like ligand small molecule or the protein with the small molecule ligand binding potential as a target point, and building a crystal structure database of targets; for these targets, collecting the information of the targets related to diseases, the biology type and the active small molecule ligand information, and integrating a comprehensive target screening database composed of an active site database, a pharmacophore database, a small molecular compound database and a target basic information database. Based on the comprehensive target screening database, the construction of the integrated drug target prediction system is realized through a script program or a PipelinePilot flow, and the probability of target prediction accuracy of the method is provided. The method provided by the invention exerts the three above technical advantages to provide the probability of target prediction, thereby providing effective basis for further experimental verification.
Owner:SICHUAN UNIV
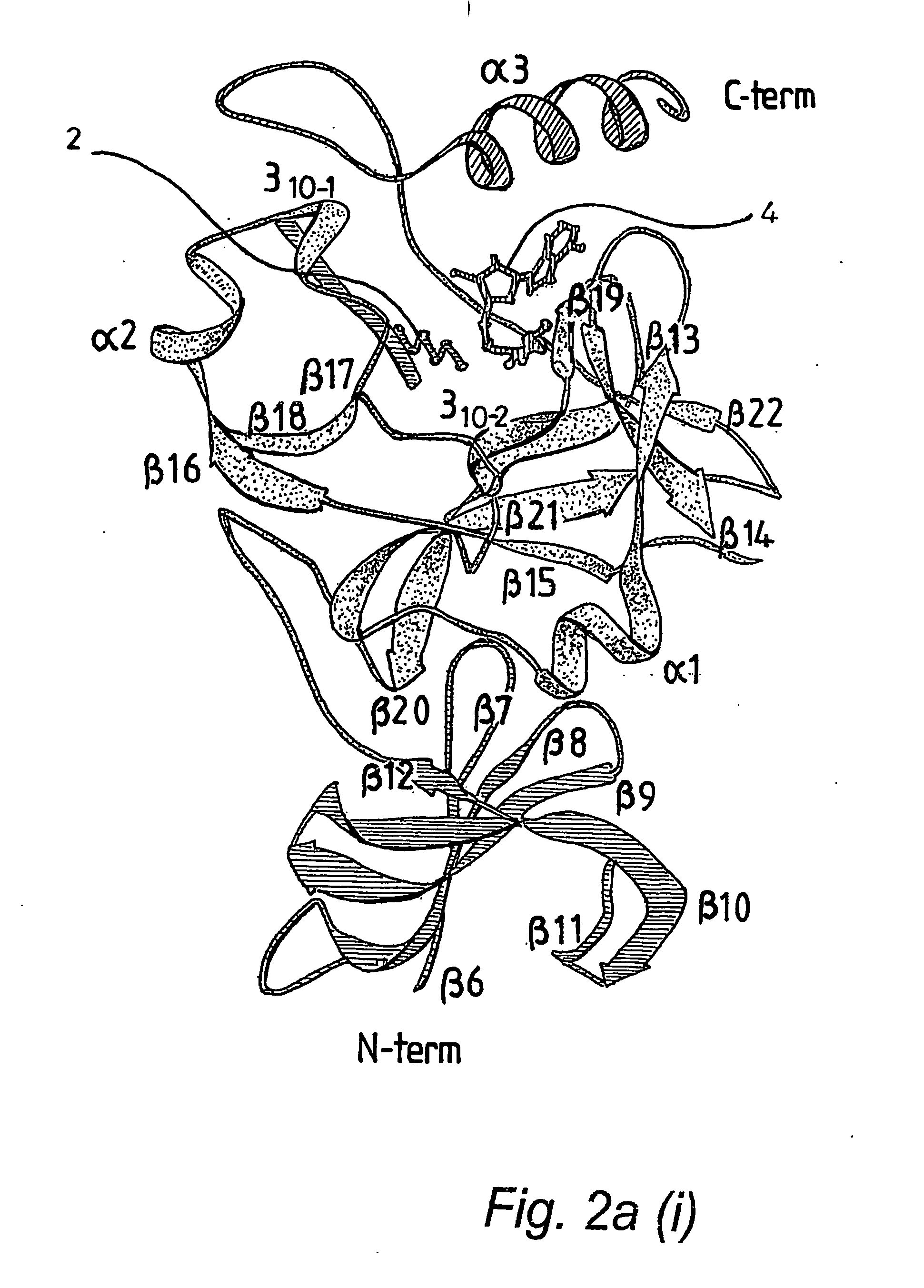
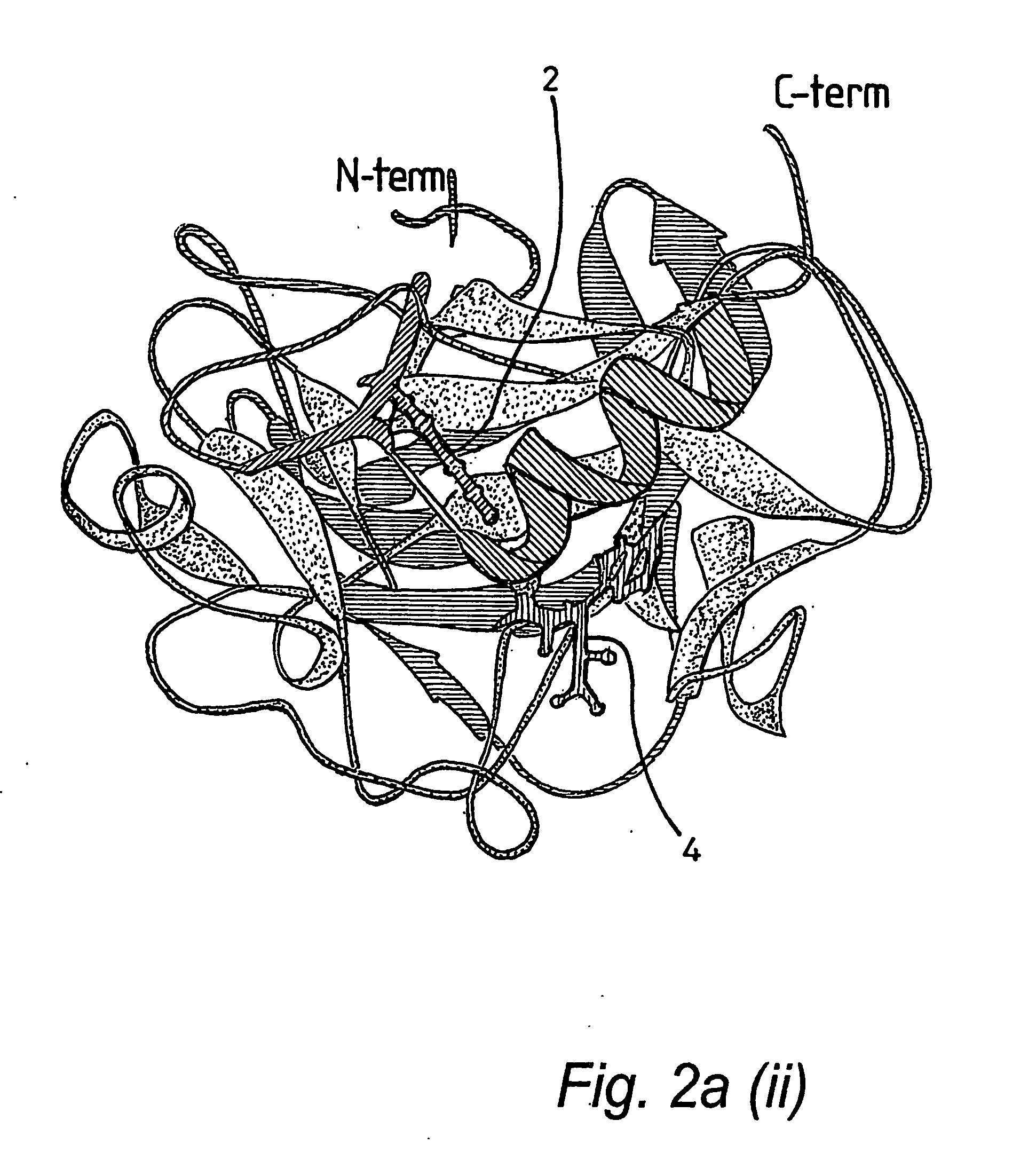
Protein crystal structure
InactiveUS20070178572A1Reduce the possibilityStrong binding specificityPolycrystalline material growthFrom normal temperature solutionsHistone methyltransferaseCrystal structure
Disclosed is a crystal comprising at least a catalytically active portion of a SET 7 / 9 histone methyltransferase, and various uses thereof, and ligands for SET 7 / 9.
Owner:MEDICAL RESEARCH COUNCIL
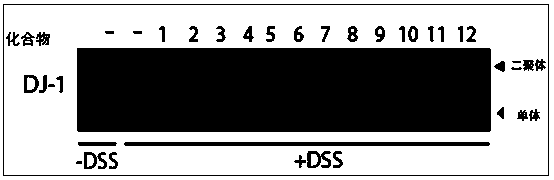
Application of active compounds for inhibiting dimerization of DJ-1
ActiveCN103768044AInhibit dimerizationGood antitumor activityAntineoplastic agentsHeterocyclic compound active ingredientsChemical compoundPharmaceutical drug
The invention provides application of active compounds in preparation of medicines for inhibiting dimerization of DJ-1. Through a computer-assisted virtualization method and technology based on a DJ-1 protein crystal structure, medicinal chemistry knowledge and pharmacological activity screening, one group of compounds for inhibiting dimerization of DJ-1 are found and the quantity of the compounds is 23. According to a principle that compounds with structure similarity coefficients Tanimoto values of more than 0.85 have similar biological activity, the molecule structure similarity Tanimoto values of more than 0.85 of the group of compounds are determined, and similar activities of inhibiting the DJ-1 are obtained. According to the application, with a DJ-1 protein relevant to the tumorigenesis development is used as a research object, compounds with functions of inhibiting the dimerization of the DJ-1 protein and inhibiting tumor cell proliferation are obtained through screening. These compounds are capable of inhibiting the dimerization of the DJ-1 protein, are strong in inhibition activity to tumor cells, and good in medicine development prospect.
Owner:ZHEJIANG UNIV
Novel methods for high efficiency and rapid getting fine three dimensional structure of target protein composite body and target molecule target molecule
ActiveCN101063682AReduce separation and purification stepsImprove bindingPreparing sample for investigationMaterial analysis using radiation diffractionNatural productActive component
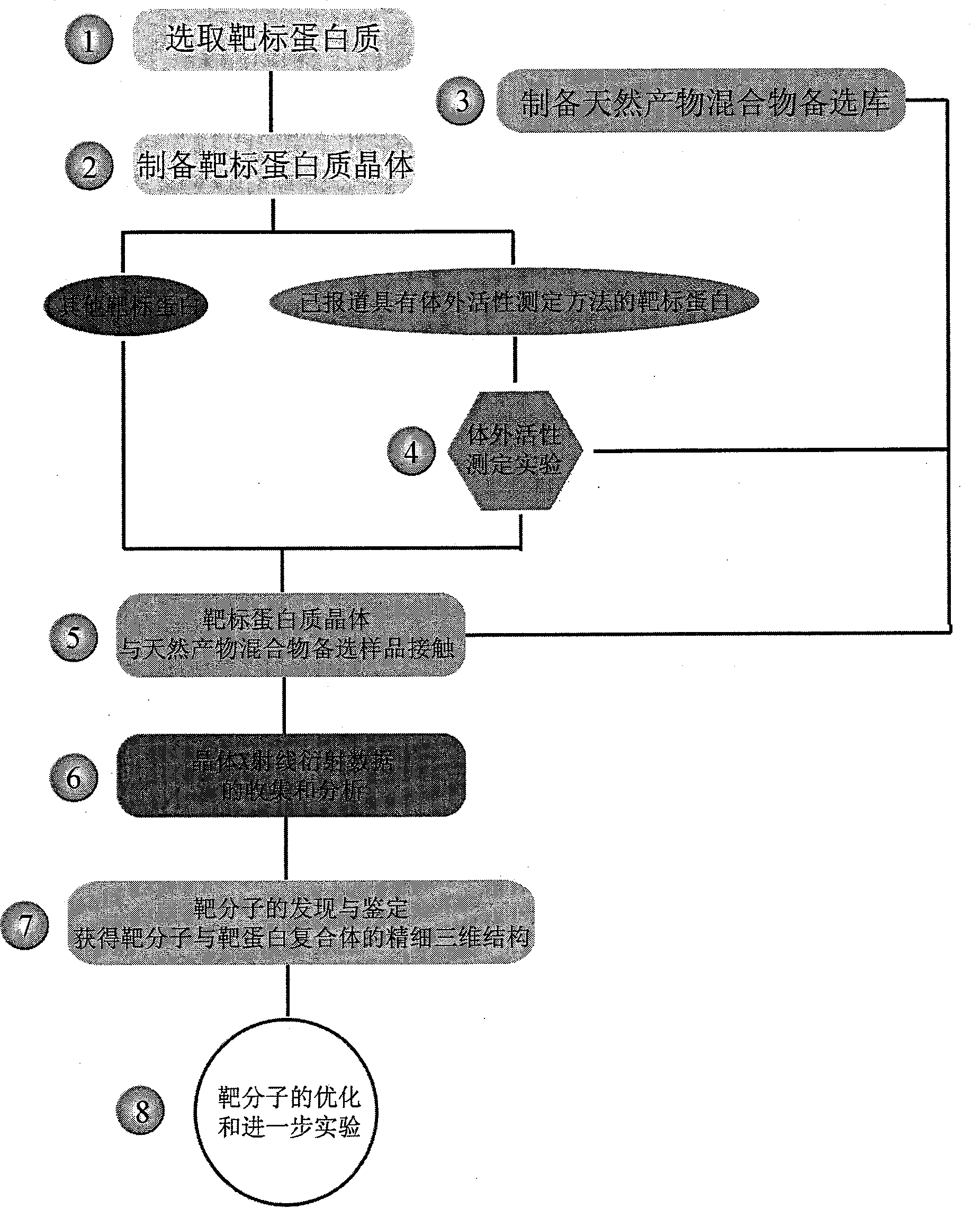
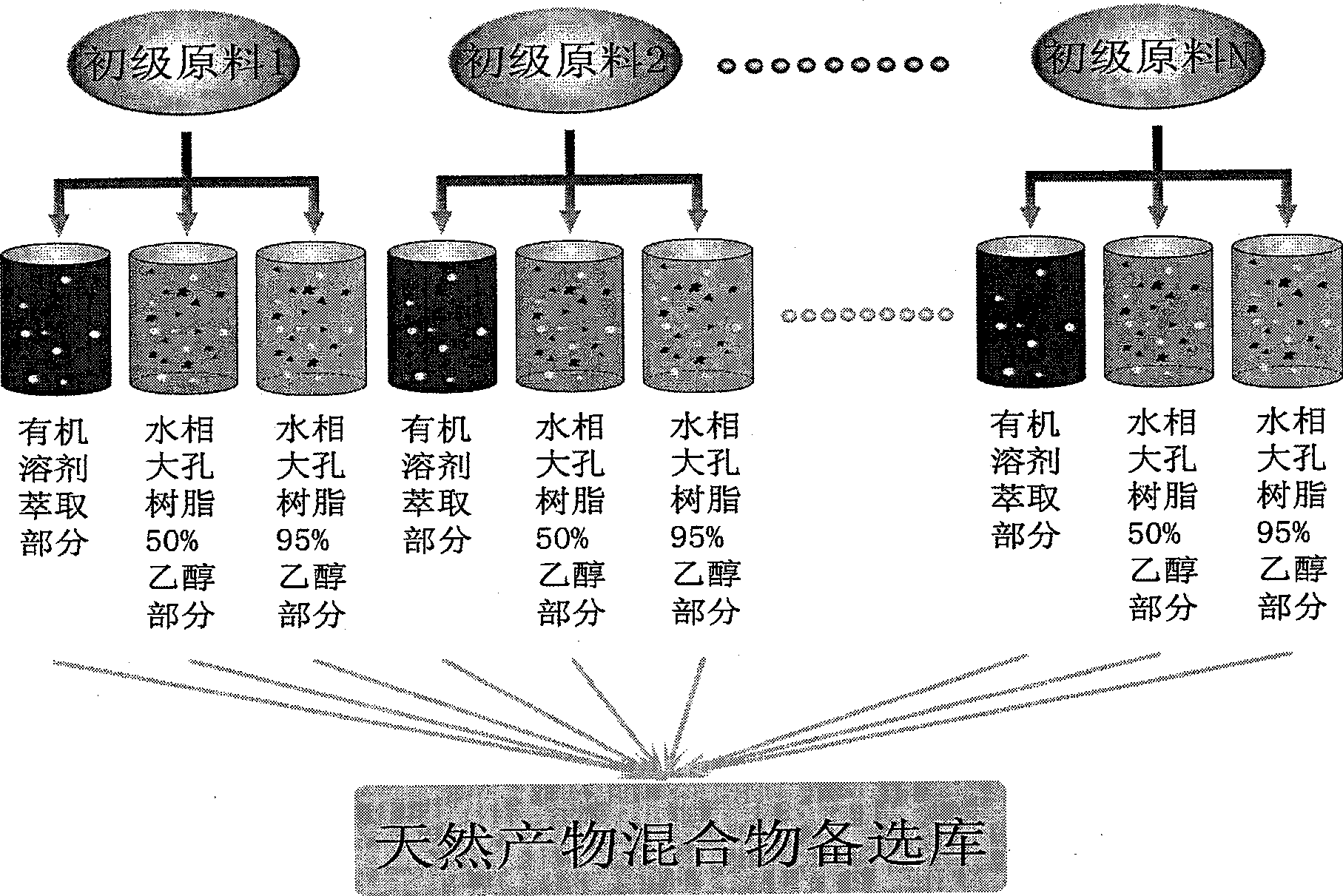
This invention provides one effective and rapid method to extract target molecule and target compound find three dimensional structure from Chinese medicine product, which uses target protein transistor stricture as base and uses X ray transistor means to filter the product for active component and for study from one new respective.
Owner:NANKAI UNIV +2
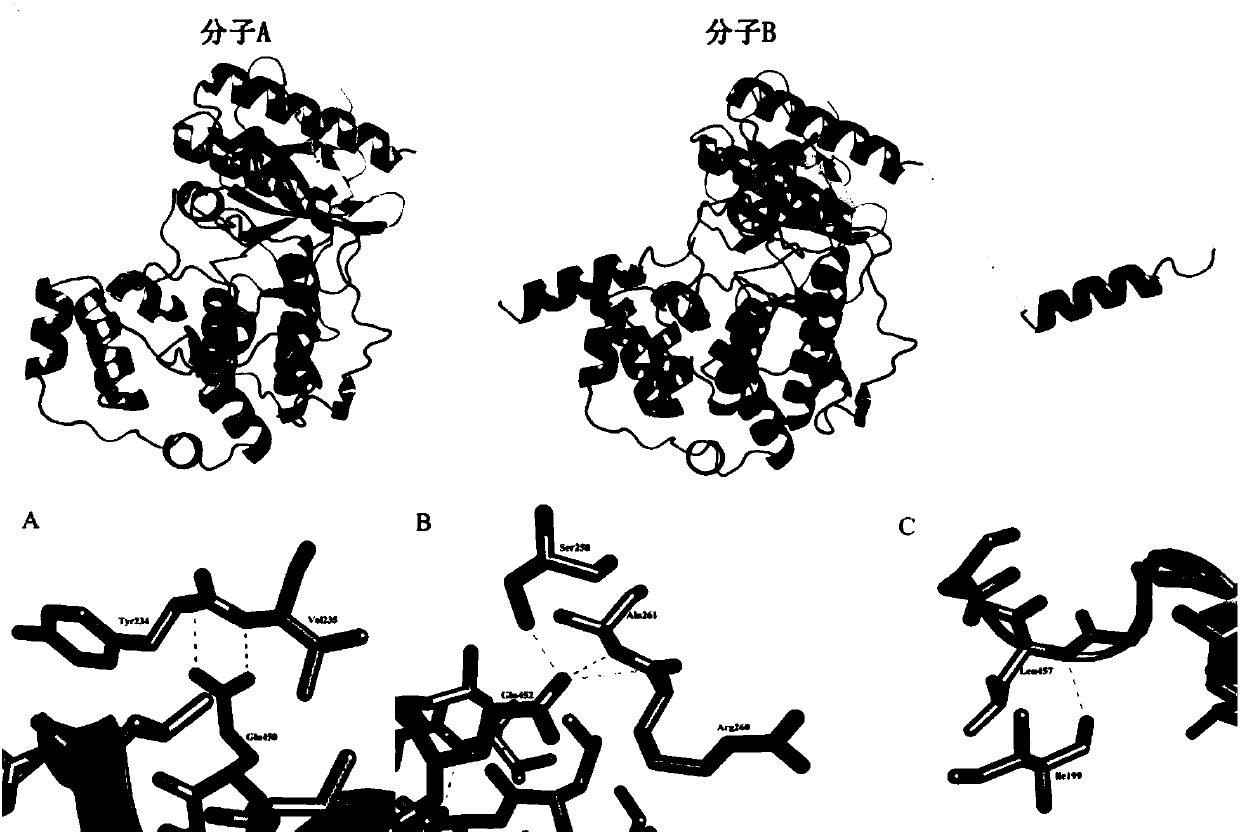
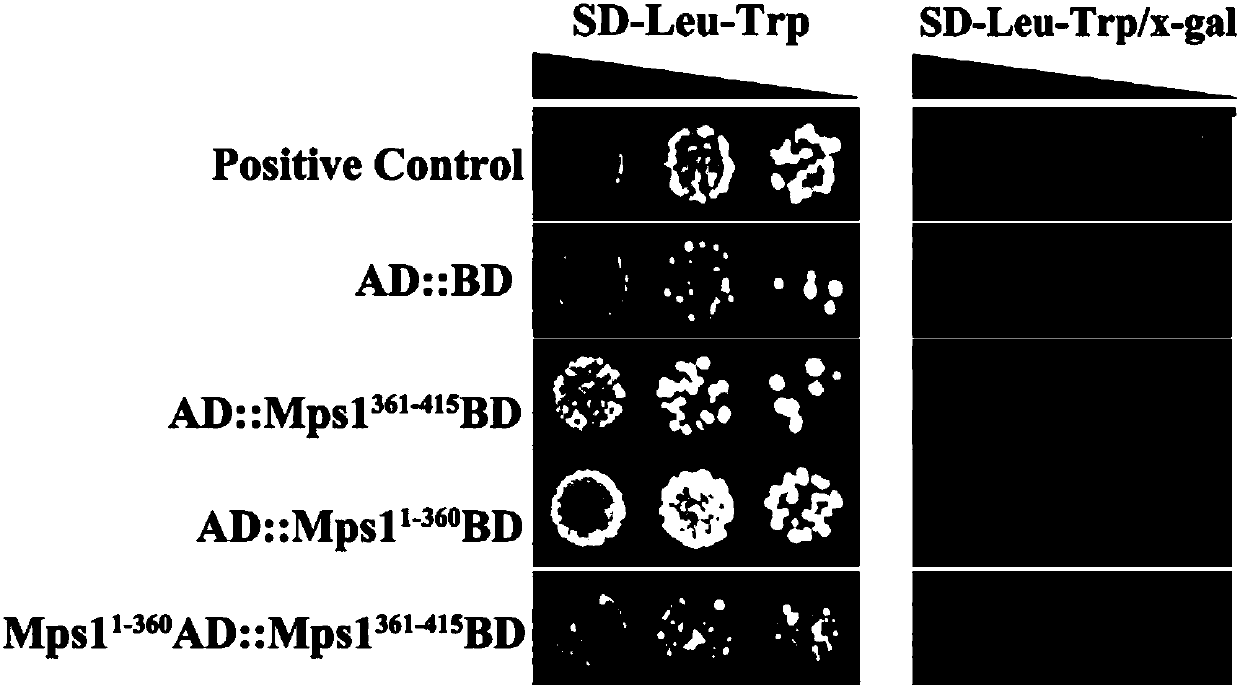
Protein crystal structure of magnaporthe oryzae mitogen-activated protein kinase Mpsl and application of protein crystal structure in bactericide target
PendingCN107904217AEnhanced inhibitory effectHigh activityTransferasesProtein structureADAMTS Proteins
The invention discloses a protein crystal structure of magnaporthe oryzae mitogen-activated protein kinase Mpsl and application of the protein crystal structure in a bactericide target. The protein crystal structure of the magnaporthe oryzae mitogen-activated protein kinase Mpsl consists of two molecules. By analysis of the protein crystal structure of the magnaporthe oryzae mitogen-activated protein kinase Mpsl, the protein crystal structure has a self-interaction effect based on structural speculateion; an experiment further verifies that the protein crystal structure has interaction betweenthe carboxyl terminal non-kinase domain (Mpsl 401-415) and the autokinase domain (Mspl 1-360), and relevant experiments verify that the carboxyl terminal non-kinase domain (Mpsl 401-415) of Mpsl hasan outstanding inhibition effect on the phosphorylation level of Mpsl, so that design, screening and optimization are performed further through the protein space structure of the carboxyl terminal non-kinase domain of Mpsl to provide a structural guidance for a small-molecular inhibitor with relatively activity of Mpsl, and a foundation is laid for development of relevant researches based on the space structure, which serves as the bactericide target, of Mpsl.
Owner:CHINA AGRI UNIV
Sorghum C2H2 zinc finger protein gene SbZFP36 and recombinant vector and expression method thereof
InactiveCN109971770AImprove stress resistanceHigh expressionPlant peptidesFermentationBiotechnologyDisease
The invention discloses a sorghum C2H2 zinc finger protein gene SbZFP36. The nucleotide sequence of the sorghum C2H2 zinc finger protein gene SbZFP36 is shown as SEQ ID NO. 1. The invention also discloses a protein encoded by the sorghum C2H2 zinc finger protein gene SbZFP36, and the amino acid sequence of the protein is shown as SEQ ID NO. 2. The invention also discloses a recombinant vector containing the sorghum C2H2 zinc finger protein gene SbZFP36 and an expression method. According to the invention, a sorghum homologous gene SbZFP36 is obtained from a sorghum genome database through theprotein sequence of a rice ZFP gene, the full length of the gene is 711bp, a prokaryotic expression vector is constructed according to the gene, a protein purification system is used for purifying theprokaryotic expression vector, the result lays a foundation for further researching the protein crystal structure and biological characteristics, and a basis is further provided for improving the disease resistance of sorghum through a gene editing method.
Owner:GUIZHOU UNIV +1
Method for screening small molecule peptidomimetic inhibitor and application of small molecule peptidomimetic inhibitor
InactiveCN111809246APotentially strong inhibitory activityThe interaction force contributes a lotLibrary screeningProteomicsVirtual screeningChemical compound
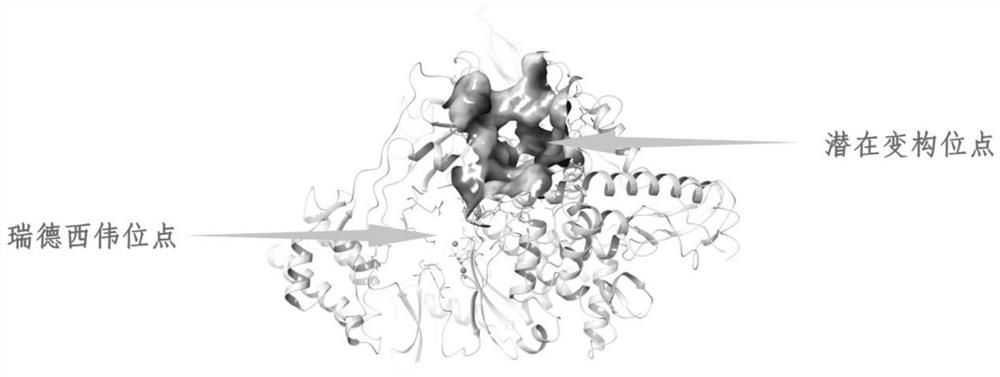
The invention discloses a method for screening a small molecular peptidomimetic inhibitor based on a COVID-19RdRp allosteric site and an application of the small molecular peptidomimetic inhibitor. The screening method specifically comprises the steps: (1) treating an SARS-Cov-2-RdRp protein crystal structure; 2) determining a drug binding site; 3) selecting a compound library; 4) treating a compound structure; 5) carrying out molecular docking; and 6) result obtaining and analysis. The advantages of virtual screening are brought into full play, the blindness of experiments is reduced, the success rate of inhibitor drugs is effectively increased, and manpower, material resources and financial resources are saved; the invention further provides the application of the inhibitor obtained through screening in preparation of drugs for treating novel coronavirus.
Owner:JINHUA VOCATIONAL TECH COLLEGE
Sorghum 14-3-3 protein GF146 gene, recombinant vector thereof and expression method
InactiveCN109943574AImprove stress resistanceHigh expressionPlant peptidesFermentationNucleotideAmino acid
The invention discloses a sorghum 14-3-3 protein GF14b gene. The nucleotide sequence of the gene is shown in SEQ ID NO.1. The invention further discloses a protein encoded by the sorghum 14-3-3 protein GF14b gene, and the amino acid sequence is shown in SEQ ID NO.2. The invention further discloses a recombinant vector containing the sorghum 14-3-3 protein GF14b gene and an expression method. Accordingly, by means of a rice GF14b gene protein sequence, a sorghum homologous gene GF14b is obtained from a sorghum genome database, and the gene is 786 BP in total length; according to the gene, a prokaryotic expression vector is constructed and purified through a protein purification system. The result lays a foundation for further studying the protein crystal structure and biological characteristics, and then the basis is provided for improving the stress resistance of sorghum through a gene editing method.
Owner:GUIZHOU UNIV
Sorghum transcription factor SbWRKY45 gene and recombinant vector containing same, and expression method of gene
InactiveCN109971767AImprove stress resistanceHigh expressionPlant peptidesFermentationBiotechnologyNucleotide
The invention discloses a sorghum transcription factor SbWRKY45 gene. A nucleotide sequence of the gene is as shown in SEQ ID NO.1. The invention further discloses a protein of a sorghum transcriptionfactor SbWRKY45 gene code, and an amino acid sequence of the protein is as shown in SEQ ID NO.2. The invention further discloses a recombinant vector containing the sorghum transcription factor SbWRKY45 gene and an expression method. A sorghum homologous gene SbWRKY45 is acquired from a sorghum gene database by a protein sequence of a rice WRKY4 gene, the overall length of the gene is 1005bp, a prokaryotic expression vector is built according to the gene, the gene is purified by a protein purification system, a base is provided for further research of a protein crystal structure and biological characteristics by a purification result, and bases are provided for improving the disease resistance of sorghum by a gene editing method.
Owner:GUIZHOU UNIV
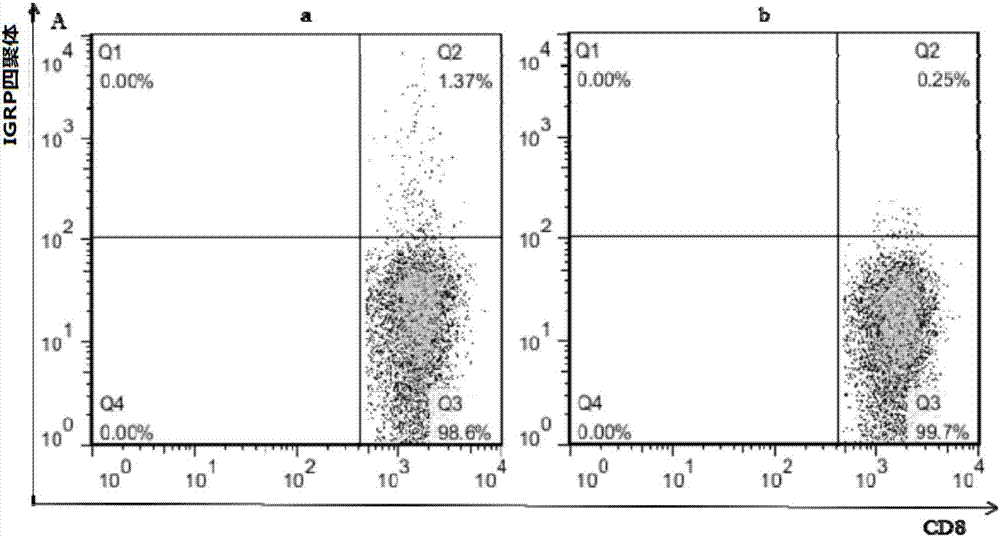
Islet-targeted protein and eukaryotic expression method thereof
InactiveCN107299092AImprove recognition rateImprove efficiencyHydrolasesVector-based foreign material introductionProtein targetMolecular binding
The invention provides an islet-targeted protein and a eukaryotic expression method thereof. The method comprises the following steps of (1) constructing plasmids containing a protein coding gene IGRP206-214-dtSCT; (2) recombining the plasmids into a chromosome of saccharomyces pastorianus; and (3) obtaining a target protein through methanol fermentation induction. A soluble IGRP206-214MHC compound is prepared through an eukaryotic expression system. Compared with the previously prepared protein for prokaryotic expression, the islet-targeted protein has galactosylated modification, and is closer to an in-vivo protein crystal structure, and the recognition rate of specific CTL on the protein and the bonding stability of TCR and an MHC molecule are improved, the endocytosis amount of viruses coupled to the protein is increased, the efficacy of the recombinant protein is improved and the recombinant protein is more stable in an external environment after galactosylated modification, and the storage period is prolonged.
Owner:李凯
Botulinum neurotoxin protein crystal structure and crystal preparation method and determination method
The invention provides a botulinum neurotoxin protein crystal structure and a crystal preparation method and determination method. The crystal has a space group P2<1>2<1>2<1>, and crystal cell parameters are as shown in the description. Through a genetic engineering method, BONT proteins being high in purity can be successfully expressed and purified, and besides, a crystallizing condition that the proteins can form perfect crystal forms can be screened; and finally, through X ray crystal diffraction, crystal data is collected, and the crystal structure of the proteins is further determined. Afoundation is provided for medicine design and an associated molecular action mechanism based on the protein structure in the future.
Owner:四川一埃科技有限公司
Polypeptide sequence bound with classical swine fever virus Erns protein and application
ActiveCN112625091AStable in natureSimple structureBiological material analysisPeptidesAssayVirus Protein
The invention mainly relates to a classical swine fever virus Erns protein affinity polypeptide and application. The polypeptide is designed by a computer-simulated virtual molecular docking technology, and the sequence is WRHYIH. According to the invention, a polypeptide sequence P1 is obtained according to a virtual molecular screening technology by taking a swine fever Erns protein crystal structure obtained by homologous modeling as a template. After the polypeptide is artificially synthesized, an enzyme-linked immunosorbent assay test and a plasma resonance test are employed to identify the affinity of the interaction of the sequence and the swine fever Erns protein, and the result shows that the sequence P1 and the swine fever Erns protein have relatively high specificity and affinity; and the invention provides a reliable theoretical basis for affinity chromatography taking affinity peptide as an affinity ligand, and swine fever Erns protein can be rapidly detected by labeling the polypeptide.
Owner:龙湖现代免疫实验室
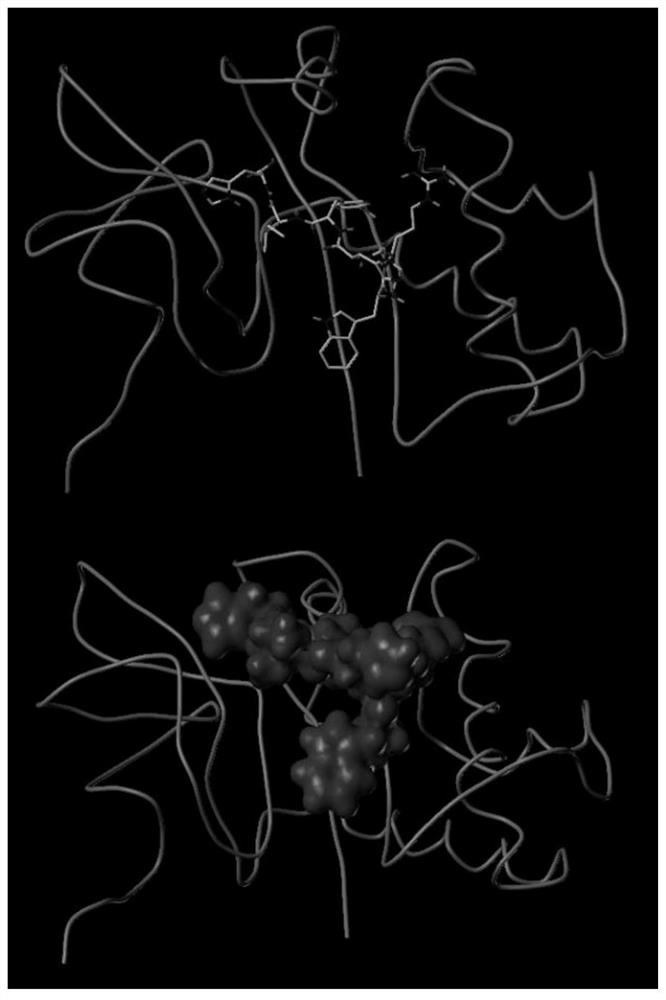
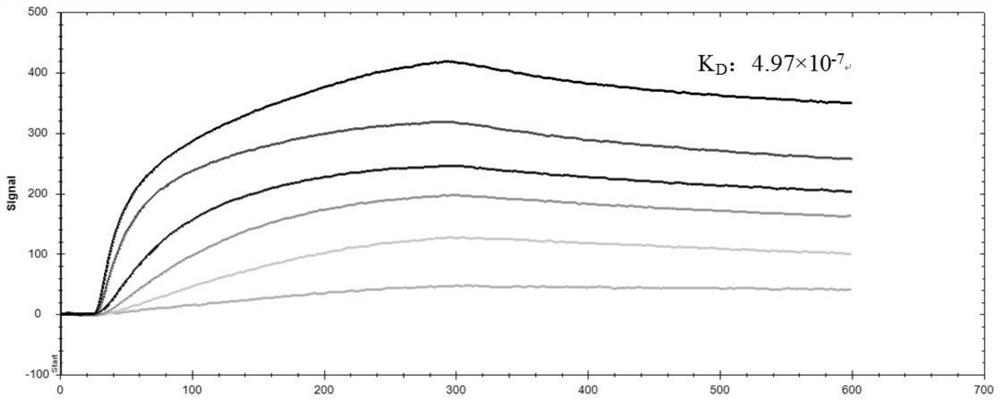
Sorghum 14-3-3 protein GF14c gene, recombined vector thereof and expression method of gene
The invention discloses a sorghum 14-3-3 protein GF14c gene. The nucleotide sequence of the gene is as shown in SEQ ID NO.1. The invention further discloses protein of a sorghum 14-3-3 protein GF14c gene code, and the amino acid sequence of the protein is as shown in SEQ ID NO.2. The invention further discloses a recombined vector and an expression method comprising the sorghum 14-3-3 protein GF14c gene. By the aid of a protein sequence of a corn GF14c gene, a sorghum homologous gene GF14c is acquired from a sorghum gene database, the overall length of the gene is 771bp, a prokaryotic expression vector is built according to the gene and purified by a protein purification system, and a foundation is laid for further study of the a protein crystal structure and biological characteristics bya purification result, so that bases are provided for improvement of sorghum stress resistance by a gene editing method.
Owner:GUIZHOU UNIV
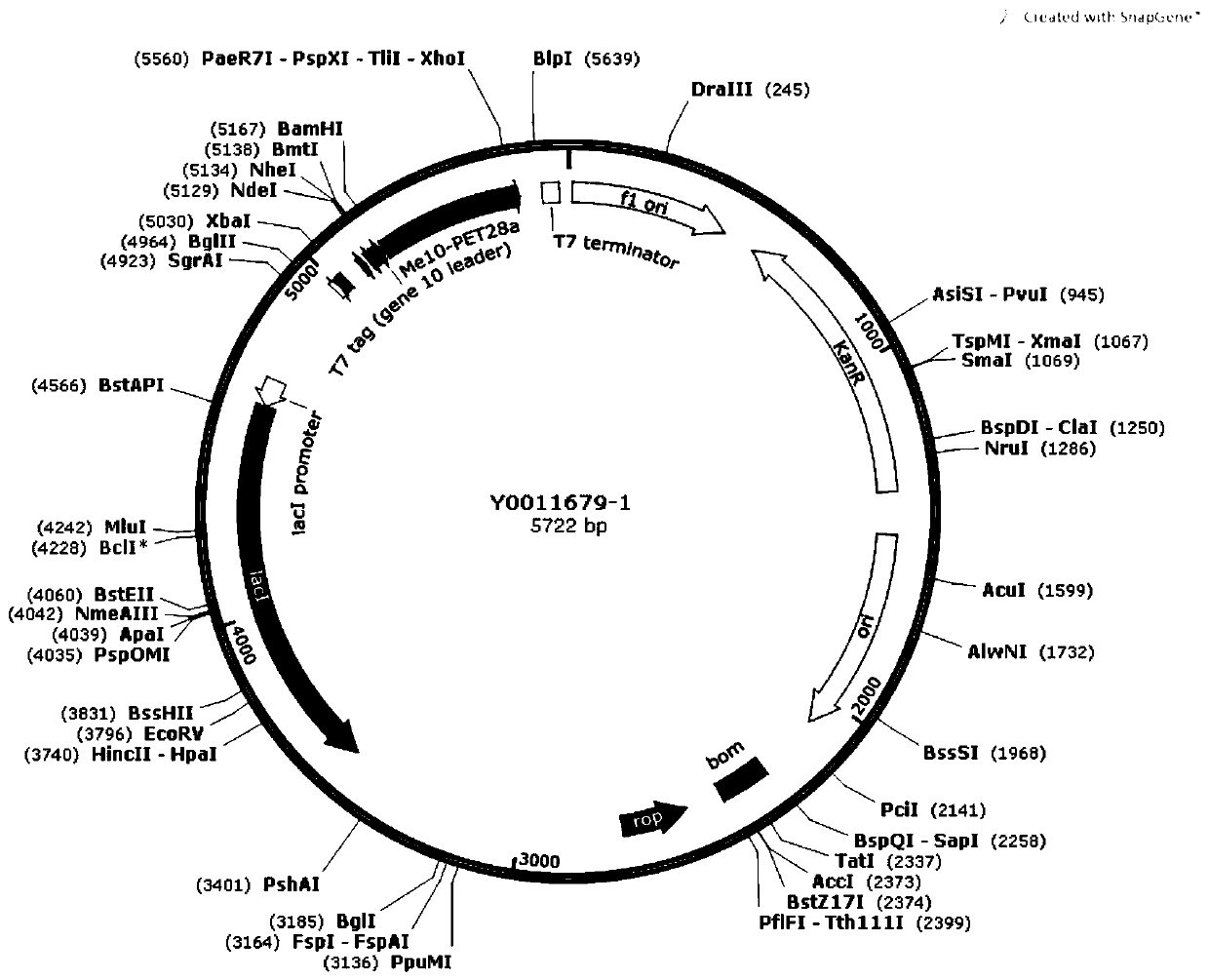
Recombinant vector and expression method of potato aphid effect protein Me10 gene
InactiveCN110117606AHigh expressionAnimals/human peptidesVector-based foreign material introductionBiotechnologyProtein structure
The invention discloses a recombinant vector of a potato aphid effect protein Me10 gene, and further discloses an expression method of the potato aphid effect protein Me10 gene. According to the recombinant vector and expression method of the potato aphid effect protein Me10 gene. The prokaryotic expression vector of the potato aphid effect protein Me10 gene is constructed, a series of optimization is carried out aiming at the protein expression condition, and finally, an Me10 soluble protein is obtained. The result lays a foundation for further study of protein crystal structures and biological characteristics, provides an idea for development of pesticides with Mp1 as a target, and then provides a basis for prevention and control of potato aphids.
Owner:GUIZHOU UNIV
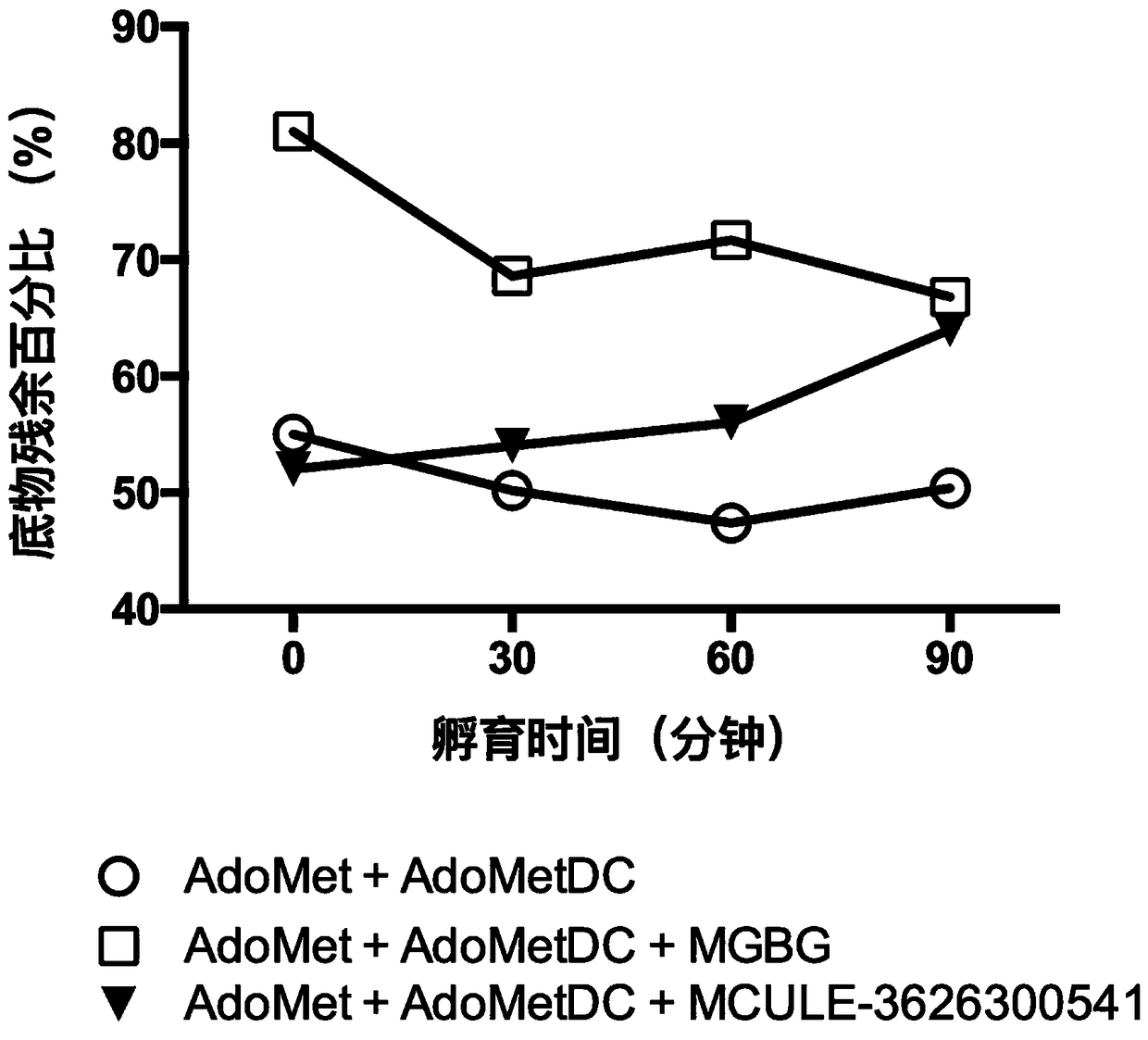
Application and Screening Method of Small Molecule Covalent Inhibitors in the Preparation of Drugs Inhibiting S-adenosylmethionine Decarboxylase
ActiveCN106389403BThe effect of decarboxylase is obviousEasy to operateOrganic active ingredientsMicrobiological testing/measurementChemical structureFuran
The invention belongs to the field of bio-medicine, and in particular relates to an application of a small-molecule covalent inhibitor in preparing a medicine for inhibiting S-adenosylmethionine decarboxylase and a screening method. The structural formula of the small-molecule covalent inhibitor is as shown in Description, wherein R1 represents phenyl or aniline or benzamide or a substituted phenyl group thereof, methyl or ethyl or furan and a substituted group thereof, pyrrole and a substituted group thereof, pyridine and a substituted group thereof and the like; and the inhibitor further consists of pharmaceutically acceptable salt with a chemical structure shown as general formula I. A method for screening the small-molecule covalent inhibitor of the S-adenosylmethionine decarboxylase comprises the following steps: deleting pyruvoyl 68 residue in an S-adenosylmethionine decarboxylase crystal structure, and conducting optimizing in Rosetta software so as to obtain an optimized protein crystal structure; and conducting docking calculation on the optimized protein crystal structure and a searched small-molecule structure by virtue of AutoDockVina software, and screening calculating results in accordance with a grading order so as to obtain the small-molecule covalent inhibitor.
Owner:CHINA THREE GORGES UNIV
Application and screening method of small molecule covalent inhibitors in the preparation of drugs for inhibiting s-adenosylmethionine decarboxylase
ActiveCN106389415BThe effect of decarboxylase is obviousEasy to operateOrganic active ingredientsComponent separationChemical structureScreening method
The invention belongs to the field of bio-medicine, and in particular relates to an application of a small-molecule covalent inhibitor in preparing a medicine for inhibiting S-adenosylmethionine decarboxylase and a screening method of the small-molecule covalent inhibitor. The structural formula of the small-molecule covalent inhibitor is as shown in Description, wherein R1 represents formyl and a derivative substituted group thereof, amino and a derivative substituted group thereof or thienyl, and R2 in an R2 group represents cyclohexene and a substituted group thereof, phenyl and a substituted group thereof, aniline or benzamide or a substituted phenyl group thereof, pyrrole and a substituted group thereof, or pyridine and a substituted group thereof; and the inhibitor further consists of pharmaceutically acceptable salt with a chemical structure shown as general formula I. A method for screening the small-molecule covalent inhibitor of the S-adenosylmethionine decarboxylase comprises the following steps: deleting pyruvoyl 68 residue in an S-adenosylmethionine decarboxylase crystal structure, and conducting optimizing in Rosetta software so as to obtain an optimized protein crystal structure; and conducting docking calculation on the optimized protein crystal structure and a searched small-molecule structure by virtue of AutoDockVina software, and screening calculating results so as to obtain the small-molecule covalent inhibitor.
Owner:CHINA THREE GORGES UNIV
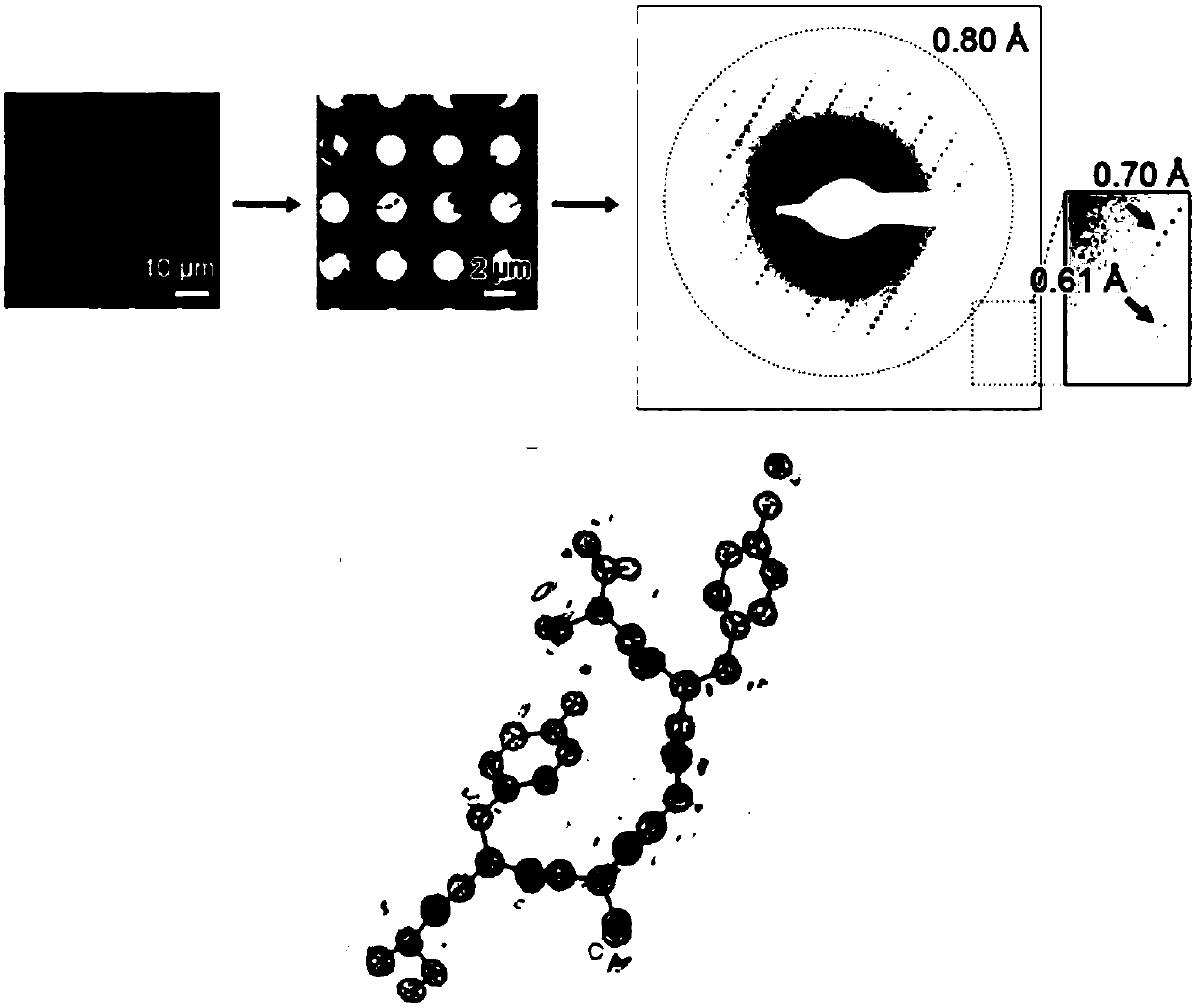
Preparation of protein micro crystal frozen sample and method of structural analysis
PendingCN110095485APreparing sample for investigationMaterial analysis using radiation diffractionImage resolutionCrystal structure
The invention provides an analytical method for the micro crystal structure of protein. Specifically, the method includes the steps of sample processing, frozen sample preparation, crystal screening,data collection, and structural analysis. According to the provided method, the structure of the protein microcrystal is analyzed by using a Cryo-SEM electron diffraction method; and the analyzed structural resolution is high and the high-resolution protein crystal structure is analyzed.
Owner:SHANGHAI INST OF ORGANIC CHEM CHINESE ACAD OF SCI
High-viscosity extrusion jet sample loading device for protein crystal structure analysis
PendingCN113588697AReduce the difficulty of buildingEasy to changeMaterial analysis using wave/particle radiationCapillary TubingCrystal structure
The invention provides a high-viscosity extrusion jet sample loading device for protein crystal structure analysis. The high-viscosity extrusion jet sample loading device comprises: a liquid inlet section connected with a hydraulic device; a piston section which is connected with the liquid inlet section and comprises a piston rod which reciprocates in the piston section; a sample section connected to the piston section, wherein the one end of the piston rod is in contact with liquid input by the liquid inlet section, and the other end of the piston rod is in contact with a viscous sample in the sample section; an air inlet section which is connected with the sample section through a capillary tube; an intersection section which is connected with the gas inlet section, wherein gas from the gas inlet section focuses the viscous sample extruded from the capillary tube into stable jet flow through shear force, and the stable jet flow is kept at the intersection section to interact with the X-ray; a recovery section which is connected with the intersection section; a fixed section which is connected with the intersection section; and an off-line separation section insertable between the sample section and the intake section. The device not only can be used for collecting crystal serial data, but also can be used for time resolution experiments of protein crystals, and has a good application prospect in the field of protein crystal structure analysis.
Owner:SHANGHAI INST OF APPLIED PHYSICS - CHINESE ACAD OF SCI
FXR-based rapid screening method for hepatotoxic compounds in polygonum multiflorum
ActiveCN113380344AEfficient screeningShorten the timeChemical property predictionMolecular entity identificationChemical structureFarnesoid X receptor
The invention relates to a method for screening hepatotoxic compounds in polygonum multiflorum based on a computer molecular docking technology. The method comprises the following steps: (1) obtaining a three-dimensional chemical structure of at least one compound to be detected in the polygonum multiflorum; (2) obtaining a protein crystal structure of the farnesoid X receptor, and determining an active site of the farnesoid X receptor; (3) respectively carrying out computer molecular docking on the compound to be detected and a positive compound with a farnesoid X receptor inhibition effect and the active sites to obtain docking scores; and (4) judging the to-be-detected compound as the hepatotoxic compound under the condition that the docking score of the to-be-detected compound reaches a docking judgment value. The screening method disclosed by the invention is rapid, economical and simple to operate, and can realize high-throughput screening of numerous compounds.
Owner:NAT INST FOR FOOD & DRUG CONTROL
Computer screening method for small molecular covalent inhibitors and application of method to screening of S-adenosylmethionine decarboxylase covalent inhibitors
ActiveCN106407739AEasy to operateGood effectSpecial data processing applicationsMolecular structuresScreening methodComputer-aided
The invention belongs to the field of biomedicine and particularly relates to a computer screening method for small molecular covalent inhibitors and an application of the method to the screening of S-adenosylmethionine decarboxylase covalent inhibitors. According to the method, in computer-aided screening and design processes of covalent inhibitors, steric hindrance of a protein receptor is reduced or eliminated first and then the protein receptor is used for screening, design and modification of the covalent inhibitors or covalent binding molecules. According to the application of the method to the screening of the S-adenosylmethionine decarboxylase covalent inhibitors, pyruvoyl 68 residues in an S-adenosylmethionine decarboxylase crystal structure are removed and optimization is performed in Rosetta software to obtain an optimized protein crystal structure; docking calculation is performed on small molecules to obtain docking calculation results; and the calculation results are screened to obtain the small molecular covalent inhibitors. According to the method and the application thereof, the perfect docking calculation method for numerous non-covalent inhibitors can be widely used, so that the screening, optimization and discovery of the covalent inhibitors can be greatly accelerated.
Owner:CHINA THREE GORGES UNIV
Application of small-molecule covalent inhibitor in preparing medicine for inhibiting S-adenosylmethionine decarboxylase and screening method of small-molecule covalent inhibitor
ActiveCN106389399AThe effect of decarboxylase is obviousEasy to operateMicrobiological testing/measurementAntineoplastic agentsFuranScreening method
The invention belongs to the field of bio-medicine, and in particular relates to an application of a small-molecule covalent inhibitor in preparing a medicine for inhibiting S-adenosylmethionine decarboxylase and a screening method of the small-molecule covalent inhibitor. The structural formula of the small-molecule covalent inhibitor is as shown in Description, wherein R1 represents phenyl or aniline or benzamide or an alkyl group or furan or pyrrole or pyridine and a substituted group thereof. A method for screening the small-molecule covalent inhibitor of the S-adenosylmethionine decarboxylase comprises the following steps: deleting pyruvoyl 68 residue in an S-adenosylmethionine decarboxylase crystal structure, and conducting optimizing in Rosetta software so as to obtain an optimized protein crystal structure; and conducting docking calculation on the optimized protein crystal structure and a searched small-molecule structure by virtue of AutoDockVina software, and screening calculating results in accordance with a grading order so as to obtain the small-molecule covalent inhibitor.
Owner:CHINA THREE GORGES UNIV
Application of a small molecule covalent inhibitor in the preparation of drugs inhibiting s-adenosylmethionine decarboxylase and its screening method
ActiveCN106389399BThe effect of decarboxylase is obviousEasy to operateMicrobiological testing/measurementAntineoplastic agentsFuranScreening method
Owner:CHINA THREE GORGES UNIV
Recombinant vector and expression method of tea tree heat shock protein CssHSP-6 gene
InactiveCN110117604AImprove disease resistanceHigh expressionPlant peptidesFermentationDiseaseProtein structure
The invention discloses a recombinant vector of a tea tree heat shock protein CssHSP-6 gene. The invention also discloses an expression method of the tea tree heat shock protein CssHSP-6 gene. A prokaryotic expression vector of the tea tree heat shock protein CssHSP-6 gene is constructed, a series of optimization is conducted on protein expression conditions, and finally a CssHSP-6 soluble proteinis obtained; the result lays a foundation for further studying the protein crystal structure and biological characteristics, and thus a basis is provided for improving the disease resistance of tea trees by a gene editing method.
Owner:GUIZHOU UNIV
Use of active compounds that inhibit dj-1 dimerization
ActiveCN103768044BInhibit dimerizationGood antitumor activityAntineoplastic agentsHeterocyclic compound active ingredientsChemical compoundPharmaceutical drug
The invention provides the application of a group of active compounds in the preparation of drugs for inhibiting DJ-1 dimerization. A group of 23 compounds that inhibit DJ-1 dimerization were discovered through computer-aided virtual methods and techniques based on DJ-1 protein crystal structure, medicinal chemistry knowledge, and pharmacological activity screening. According to the compounds whose structural similarity coefficient Tanimoto value is greater than 0.85, have similar biological activity principles, it is determined that the molecular structural similarity Tanimoto value of this group of compounds is greater than 0.85, and also have similar inhibitory DJ-1 activity. The invention takes the DJ-1 protein related to the occurrence and development of tumor as the research object, and screens to obtain the compound capable of inhibiting the dimerization of the DJ-1 protein and inhibiting the proliferation of tumor cells. These compounds can not only inhibit the dimerization of DJ-1 protein, but also have strong inhibitory activity on tumor cells, and have good prospects for drug development.
Owner:ZHEJIANG UNIV
Chemical reaction parameter calculation method based on computer
PendingCN114334023AUnderstand natureChemical processes analysis/designComputational theoretical chemistryChemical reactionProtein structure
The invention discloses a computer-based chemical reaction parameter calculation method, which comprises the following steps of: S1, collecting an initial structure of protein in a protein crystal structure file, and obtaining an initial structure file of catalytic enzyme on the basis of the protein crystal structure file; s2, importing the protein initial structure file into AMBER software, performing molecular dynamics simulation, and simulating a protein environment; s3, recording the change of the protein structure under different interference conditions by taking different external environments (such as different pH values and the like) as the interference conditions; s4, importing the protein structure file subjected to molecular dynamics simulation into Gaussian software; and optimizing the structure by using Gaussian software. According to the chemical reaction parameter calculation method based on the computer, the amino acid and substrate effects of the catalytic enzyme in different states are obtained through rapid calculation capability, so that people can better understand the properties of various catalytic enzymes, and later application is facilitated.
Owner:INNER MONGOLIA UNIV OF TECH
A novel method for obtaining fine three dimensional structures of target molecules and target protein composite bodies efficiently and rapidly from a mixing system of natural products of traditional C
ActiveCN100465643CReduce separation and purification stepsReduce soakingPreparing sample for investigationMaterial analysis using radiation diffractionProtein targetNatural product
This invention provides one effective and rapid method to extract target molecule and target compound find three dimensional structure from Chinese medicine product, which uses target protein transistor stricture as base and uses X ray transistor means to filter the product for active component and for study from one new respective.
Owner:NANKAI UNIV +2
Polypeptide capable of specifically binding with human vascular endothelial growth factor receptor-3 (VEGFR-3) protein and screening method and identification and application of polypeptide
InactiveCN105693821AEfficient screeningHigh affinityEnergy modified materialsPeptidesTumor targetProtein target
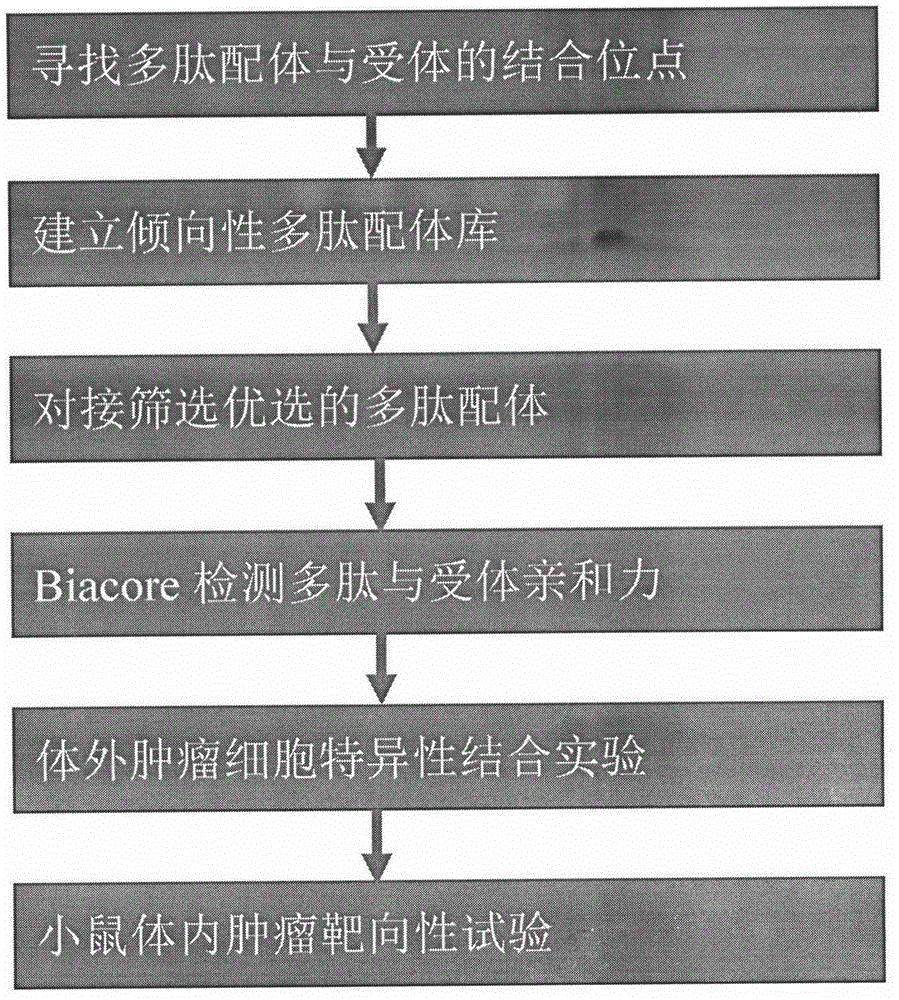
Based on the protein crystal structure of human vascular endothelial growth factor receptor-3 (VEGFR-3), the present invention designs and develops a polypeptide that specifically binds to it, and discloses its screening method, identification and use. The general formula of the amino acid sequence of the polypeptide is CXXPFXP, X is any L-type or D-type amino acid in 20 kinds of natural amino acids. The present invention also relates to the amide, ester or salt of the polypeptide, or the active fragment, active derivative and fusion polypeptide formed by the polypeptide, or the polymer formed by using the polypeptide as a ligand. Using computer simulation screening and immunofluorescence in vitro verification to obtain tumor-targeting polypeptides with high expression of VEGFR-3, it is convenient and quick. The screened polypeptides have clear structures, high affinity with target proteins, almost no immunogenicity, and are easy to prepare and produce. It has broad application prospects in the fields of tumor-targeted drug delivery, imaging and diagnosis, and gene transfection.
Owner:CHINA PHARM UNIV
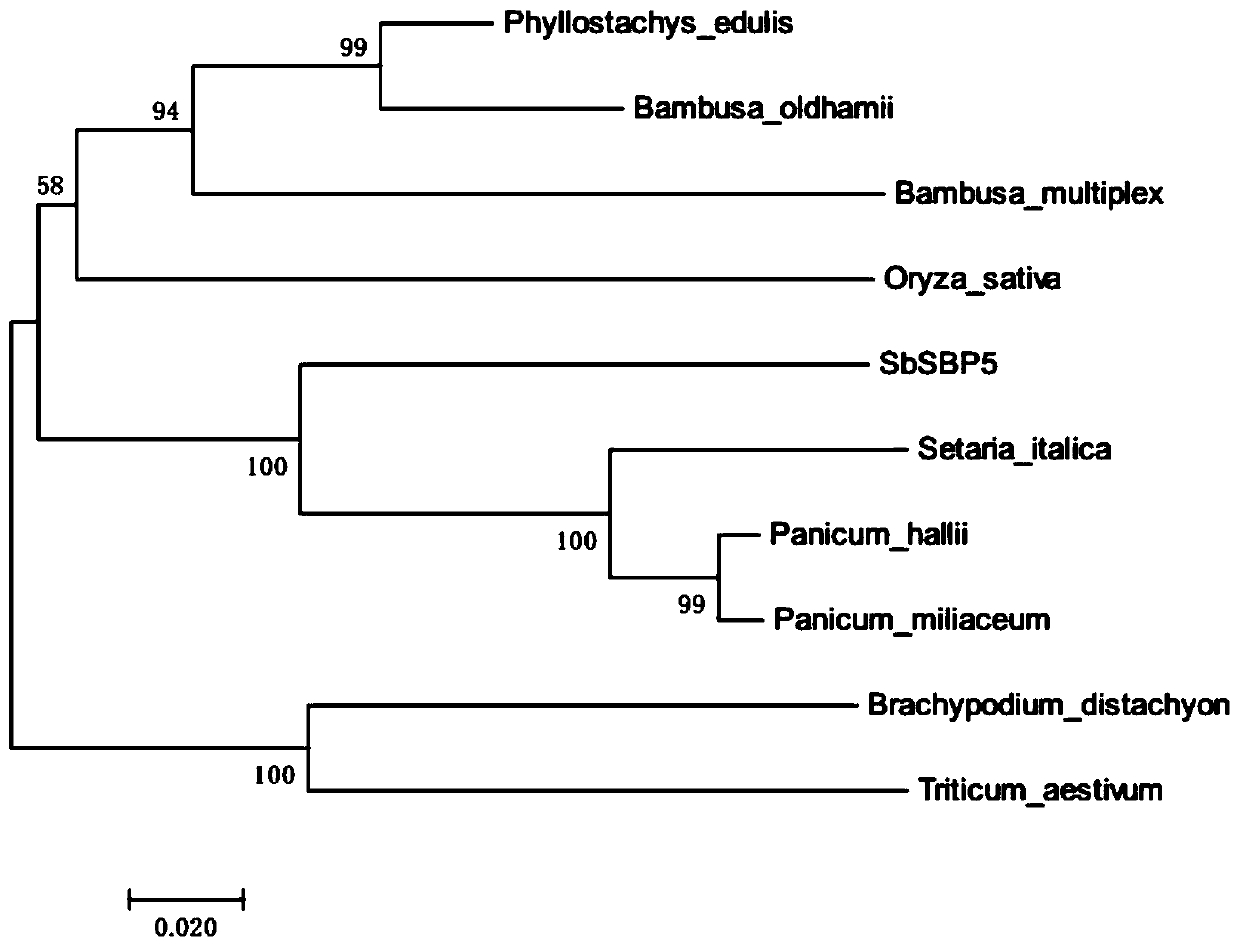
Sorghum transcription factor SbSBP5 gene and recombinant vector and expression method thereof
InactiveCN109971768AImprove stress resistanceHigh expressionPlant peptidesFermentationBiotechnologyNucleotide
The invention discloses a sorghum transcription factor SbSBP5 gene, and the nucleotide sequence of the sorghum transcription factor SbSBP5 gene is shown as SEQ ID NO. 1. The invention also discloses aprotein encoded by the sorghum transcription factor SbSBP5 gene, and the amino acid sequence of the protein is shown as SEQ ID NO. 2. The invention further discloses a recombinant vector containing the sorghum transcription factor SbSBP5 gene and an expression method. According to the invention, a sorghum homologous gene SbSBP5 is obtained from a sorghum genome database through the protein sequence of a rice SBP gene, the full length of the gene is 1227bp, a prokaryotic expression vector is constructed according to the gene, a protein purification system is used for purifying the prokaryoticexpression vector, the result lays a foundation for further researching a protein crystal structure and biological characteristics, and a basis is provided for improving the disease resistance of sorghum through a gene editing method.
Owner:GUIZHOU UNIV +1
Sorghum drought response gene SbJAZ1 and recombinant vector and expression method thereof
PendingCN109971769AImprove stress resistanceHigh expressionPlant peptidesFermentationBiotechnologyNucleotide
The invention discloses a sorghum drought response gene SbJAZ1. The nucleotide sequence of the sorghum drought response gene SbJAZ1 is shown as SEQ ID NO. 1. The invention also discloses a protein encoded by the sorghum drought response gene SbJAZ1, and the amino acid sequence of the protein is shown as SEQ ID NO. 2. The invention further discloses a recombinant vector containing the sorghum drought response gene SbJAZ1 and an expression method. According to the method, a sorghum homologous gene SbJAZ1 is obtained from a sorghum genome database through the protein sequence of a rice JAZ1 gene,the full length of the gene is 606bp, a prokaryotic expression vector is constructed according to the gene, the condition of soluble expression of the gene is optimized, meanwhile, the protein purification system is used for purifying the gene, the result lays a foundation for further researching the protein crystal structure and biological characteristics, and a basis for further provided for improving the drought resistance of sorghum through a gene editing method.
Owner:GUIZHOU UNIV +1
Construction and prediction method of integrated drug target prediction system
InactiveCN102663214BOvercoming the problem of low forecasting accuracySpecial data processing applicationsPharmacophoreEngineering
Owner:SICHUAN UNIV
Features
- R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
Why Patsnap Eureka
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Social media
Patsnap Eureka Blog
Learn More Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com