Computer screening method for small molecular covalent inhibitors and application of method to screening of S-adenosylmethionine decarboxylase covalent inhibitors
A technology of covalent inhibition of adenosylmethionine decarboxylase, which is applied in the field of biomedicine, can solve problems such as tail deviation, and achieve the effects of accelerated screening, simple operation, and good predictive effect
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
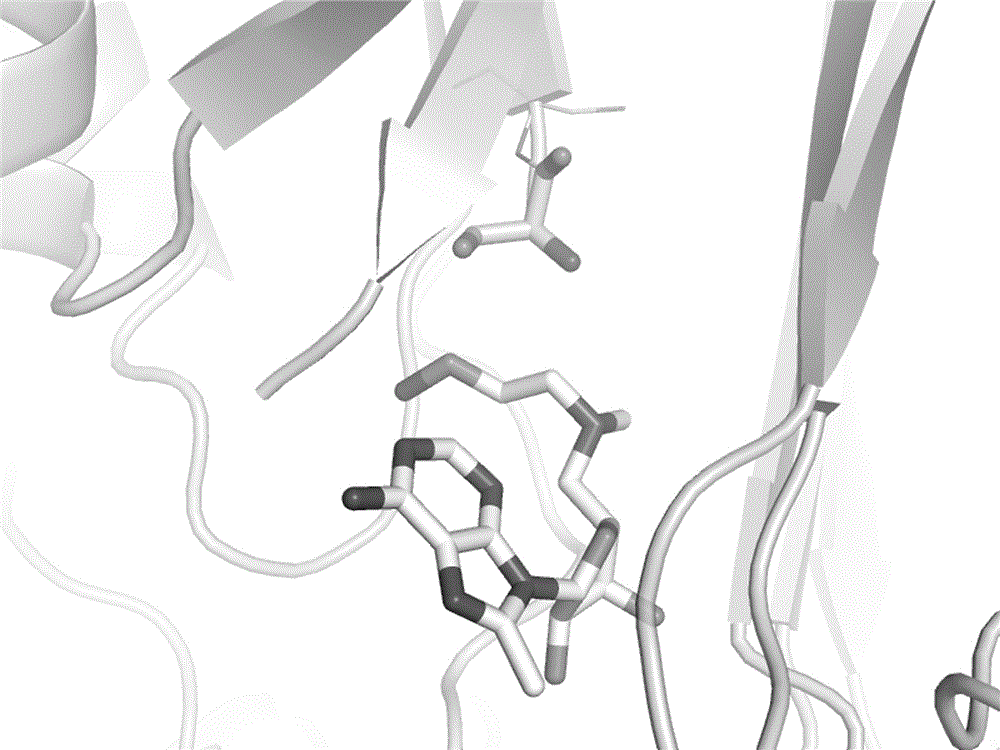
[0047] 1) Delete the pyruvyl 68 (Pyr) residue in the crystal structure of S-adenosylmethionine decarboxylase (AdoMetDC), and optimize it in Rosetta to obtain the SCAR protein of AdoMetDC;
[0048] 2) Use AutoDockVina to carry out the docking calculation of covalent compounds, and dock the corresponding AdoMetDC covalent inhibitor molecule in the PDB code 3DZ5 into the SCAR protein structure of AdoMetDC, the conformation is as shown in the figure figure 2 As shown, the docked conformation matches well with the crystal structure conformation.
Embodiment 2
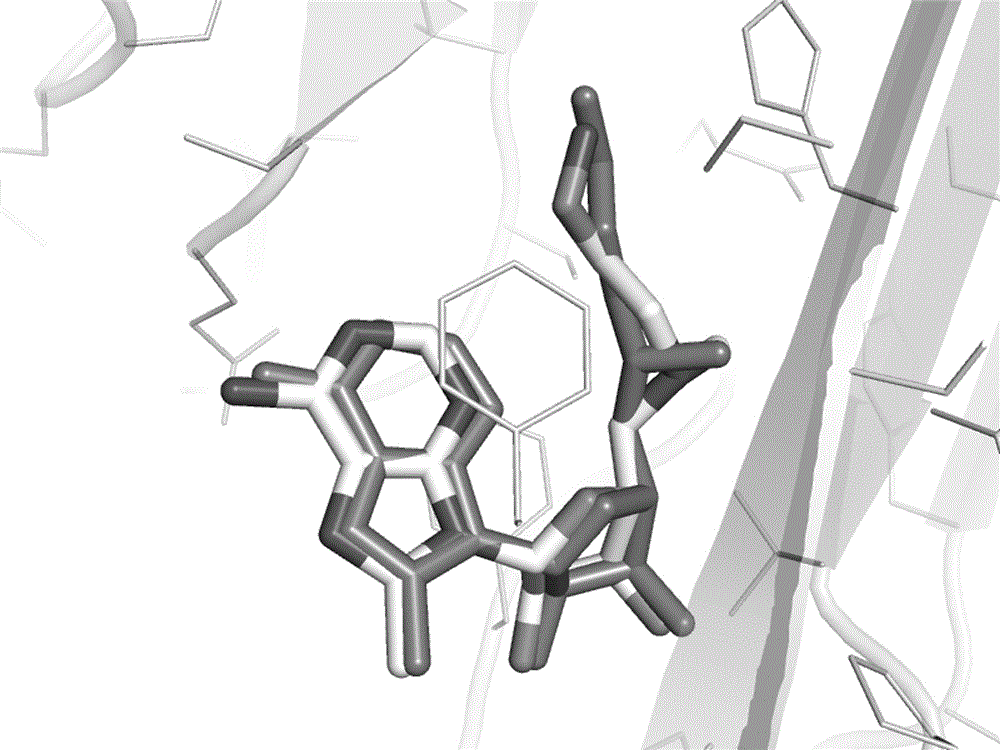
[0050] 1) Delete the pyruvyl (Pyr) 68 residue in the crystal structure of S-adenosylmethionine decarboxylase (AdoMetDC), and optimize it in Rosetta to obtain the SCAR protein of AdoMetDC;
[0051] 2) Use AutoDockVina to carry out the docking calculation of covalent compounds, and dock the corresponding AdoMetDC covalent inhibitor molecule in PDB code 1I72 into the SCAR protein structure of AdoMetDC, the conformation is as shown in the figure image 3 As shown, the docked conformation matches well with the crystal structure conformation.
Embodiment 3
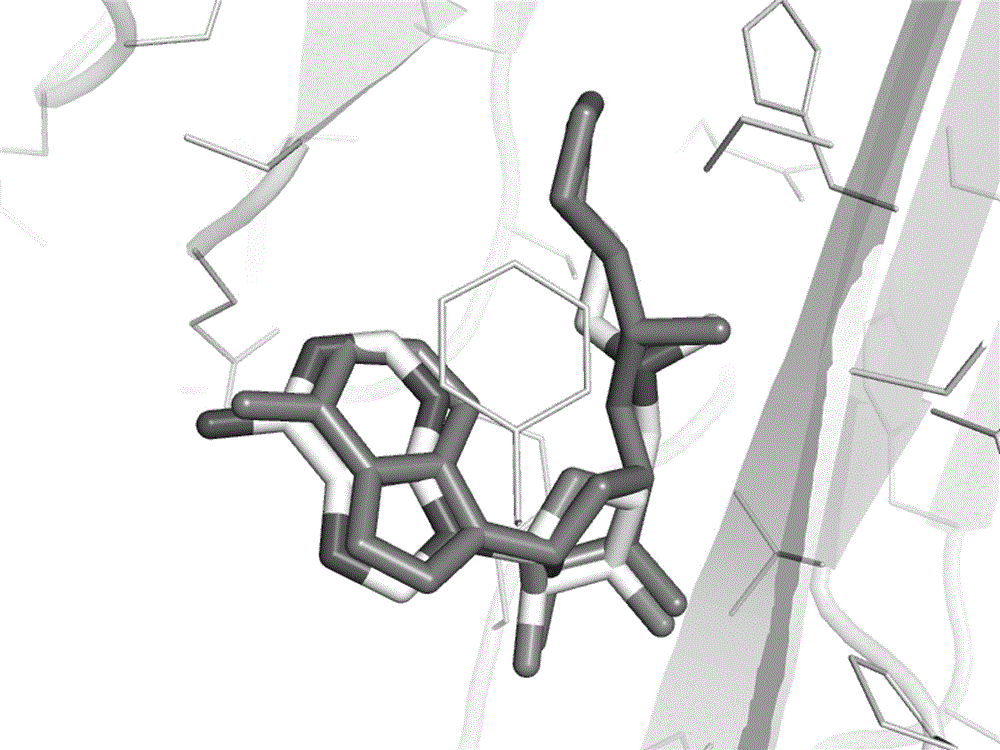
[0053] 1) Delete the pyruvyl (Pyr) 68 residue in the crystal structure of S-adenosylmethionine decarboxylase (AdoMetDC), and optimize it in Rosetta to obtain the SCAR protein of AdoMetDC;
[0054] 2) Use AutoDockVina to carry out the docking calculation of covalent compounds, and dock the AdoMetDC covalent inhibitor molecule corresponding to the PDB code 1I79 into the SCAR protein structure of AdoMetDC. The conformation is as shown in the figure Figure 4 As shown, the docked conformation matches well with the crystal structure conformation.
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com