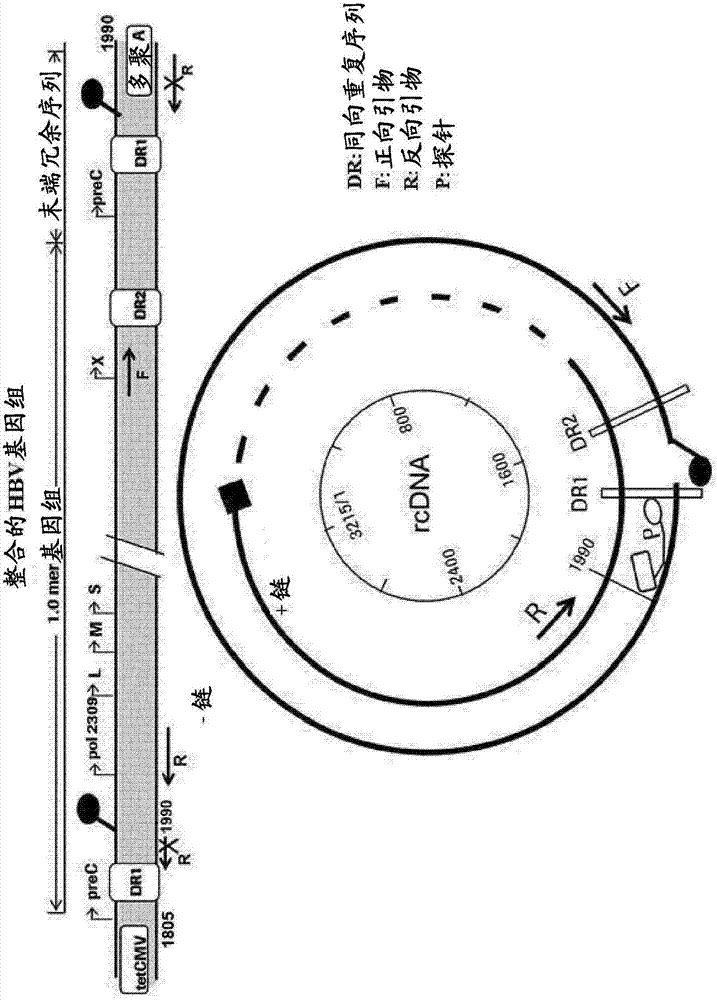
A novel high-throughput method for quantification of HBV cccDNA from cell lysate by real-time PCR
A primer pair, reverse primer technology, applied in the high-throughput field
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
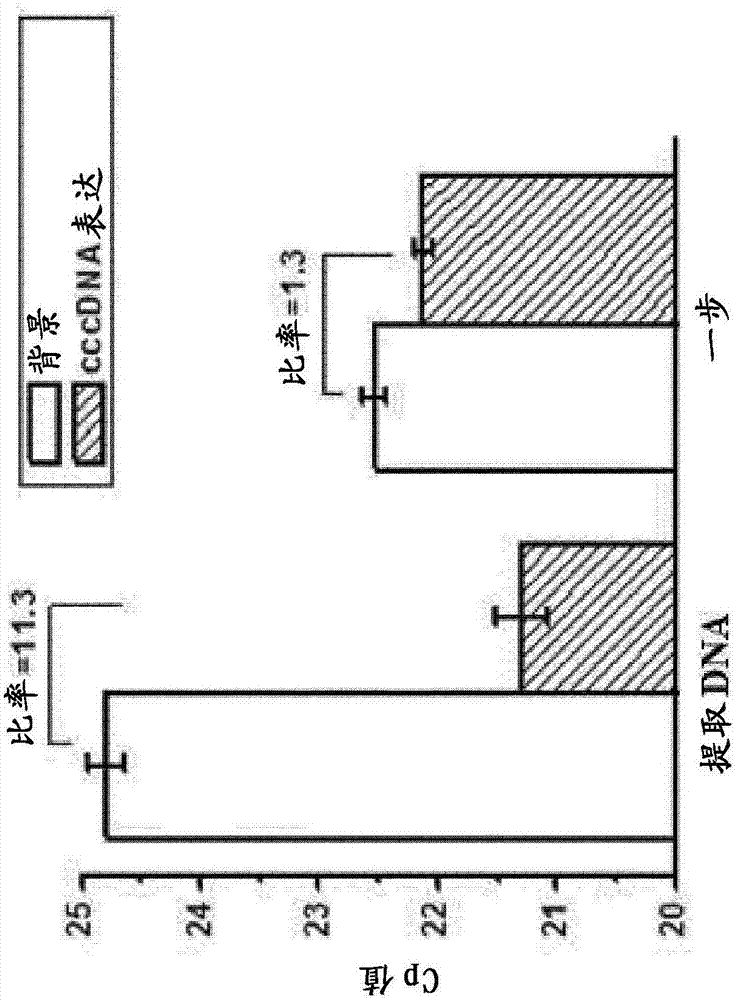
[0135] Example 1: Comparison of a qPCR assay using a conventional cccDNA primer pair against extracted viral genomic DNA and a one-step qPCR assay
[0136] The source of cell material was HepDES19 with no induction of cccDNA (background, due to the presence of 1 μg / mL tetracycline) or induction (cccDNA expression, due to the absence of tetracycline) after six days of culture. Test the regular group. DNA extraction methods and one-step qPCR assays were performed following the procedures described above.
[0137] figure 2 The results shown in describe the range between DNA from Tet-on (background) conditions and DNA from Tet-off (cccDNA expression) conditions. For the extracted DNA method, the conventional group showed an 11.3-fold difference in the qPCR assay. For the one-step qPCR assay, there was only a 1.3-fold difference. By comparing the results of the extracted DNA method and the one-step qPCR assay, the conventional panel could not be used to establish a one-step cc...
Embodiment 2
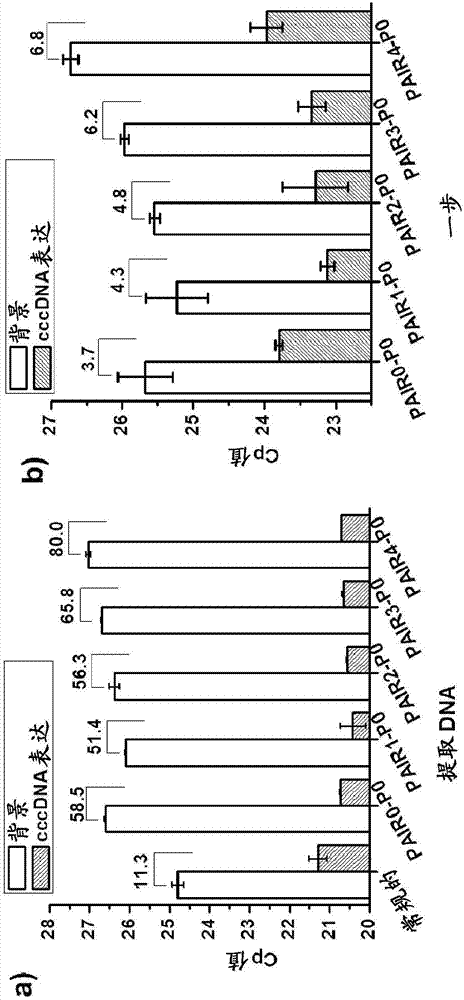
[0138] Example 2: Test of new cccDNA primers for detection of extracted DNA from HepDES19 or direct lysate without extraction (one-step qPCR assay)
[0139]The source of cell material was HepDES19 with no induction of cccDNA (background, due to the presence of 1 μg / mL tetracycline) or induction (cccDNA expression, due to the absence of tetracycline) after six days of culture. The following materials were used in this example: conventional set, PAIR0-P0, PAIR1-P0, PAIR2-P0, PAIR3-P0 and PAIR4-P0. The dye used in P0 was TAMRA and the quencher used in P0 was BHQ2. A one-step qPCR assay was performed following the procedure described above.
[0140] image 3 The results shown in a show that the range between extracted DNA from background and extracted DNA from cccDNA expression in HepDES19 cells was improved from 11.3-fold in conventional group to PAIR0-P0, PAIR1-P0, PAIR2-P0, PAIR3- 50-80 times in P0 and PAIR4-P0. This indicates that the new primer pairs and probes, namely PA...
Embodiment 3
[0141] Example 3: Experimentation of new cccDNA probes for detection of extracted DNA from HepDES19 or direct lysates of a one-step qPCR assay without extraction (one-step qPCR assay).
[0142] Cell material was derived from HepDES19 with no cccDNA induction (background, due to the presence of 1 μg / mL tetracycline) or induction (cccDNA expression, due to the absence of tetracycline) after six days of culture.
[0143] For DNA extraction studies, the probes used were P0, P1, P2 and P3, where in each probe the dye used was TAMRA and the quencher BHQ2 was used. The primer used for all reactions was PAIR4.
[0144] For the one-step qPCR assay, the probe used was P2, and the dye used in P2 was TAMRA and the quencher used in P2 was BHQ2. The primers used were PAIR1 and PAIR4, respectively. A conventional group was also used for control.
[0145] Figure 4 The results shown in a describe the design of new probes and a comparison of their ability to detect HBVcccDNA in extracted v...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com