Large-batch unicellular ATAC-seq data quality controlling and analyzing method
A quality control method and data quality technology, applied in the field of biology, can solve problems such as the development of a quality control analysis process for single-cell epigenome sequencing data
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment Construction
[0024] In order to understand the technical content of the present invention more clearly, the following examples are given in detail.
[0025] Hereinafter, the present invention will be more fully described and illustrated by using exemplary embodiments of the present invention with reference to the accompanying drawings, but it does not mean that the present invention is limited thereto.
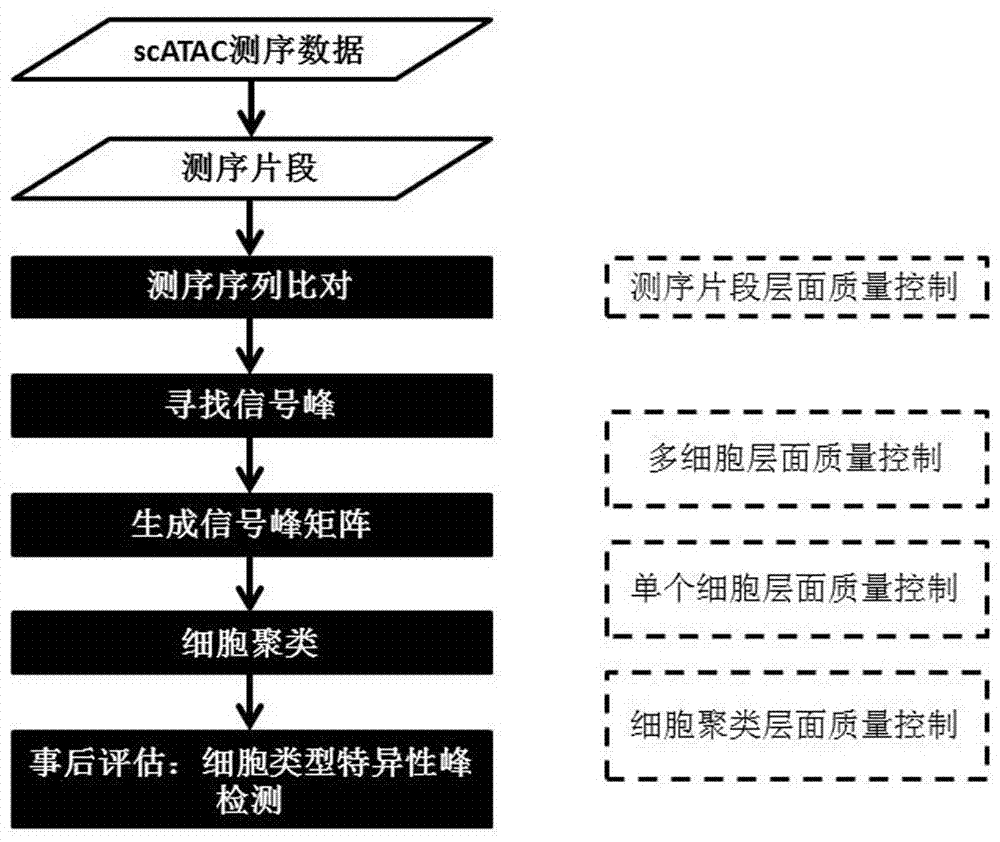
[0026] Such as figure 1 Shown is a flow chart of the steps of the scATAC-seq data quality control and data analysis process control method of the present invention.
[0027] In one embodiment, the quality control and analysis method uses the scATAC-seq data set, the data comes from the NCBI GEO database (GSE65360), and the data comes from a total of 288 scATAC-seq data sets of three cell types combined into one set scATAC-seq data. Such as figure 1 shown, including the following steps:
[0028] The first step is to process the data file: the FASTQ format of the original sequencing fi...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com