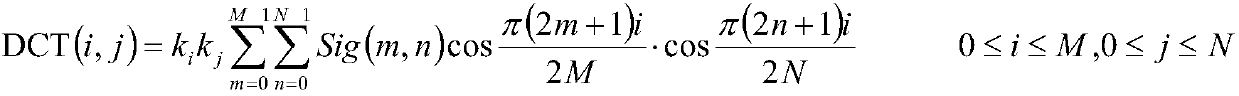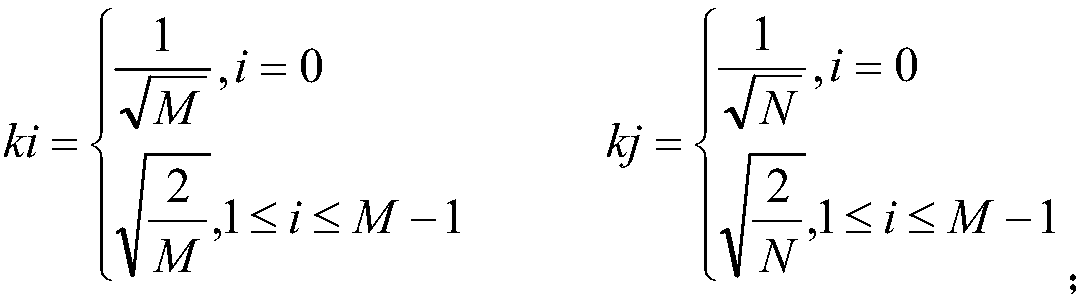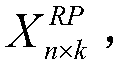Method for determining interaction between proteins based on random projection integrated classification
A technology of random projection and measurement method, applied in the field of biology, can solve the problems of not meeting the technical development requirements, high false positive rate, low efficiency, etc., and achieve the effects of excellent measurement stability, precise expression, and excellent accuracy
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment Construction
[0028] A method for determining the interaction between proteins based on random projection integrated classification, characterized in that it comprises the following steps:
[0029] A. Protein data screening
[0030] Screen protein interaction pairs from the protein database DIP;
[0031] B. Substitution Matrix Representation
[0032] Using BLOSUM62 matrix, a protein sequence of length N will generate an N×20 matrix, and the SMR matrix represents the expression: SMR(i,j)=B(P(i),j) i=1...N,j =1...20,
[0033] B(P(i), j) represents the probability that amino acid i is mutated into amino acid j, and P(i), j represents the protein sequence position composed of N amino acids;
[0034] C. Discrete cosine transform
[0035] The discrete cosine transform DCT formula is as follows:
[0036]
[0037] in,
[0038] D. Establish a random projection ensemble model
[0039] Select the original matrix X of n×d dimensions n×d , the original matrix is mapped to obtain a low-dim...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com