A Universal Primer and Its Design and Amplification Method for Transcriptional Spacer 2 in Ribosome Ribosome of Pleuriformes Soleidae Fish
An internal transcribed spacer, universal primer technology, applied in biochemical equipment and methods, microbial determination/inspection, DNA/RNA fragments, etc. The effect of obvious difference, efficient and quick identification
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
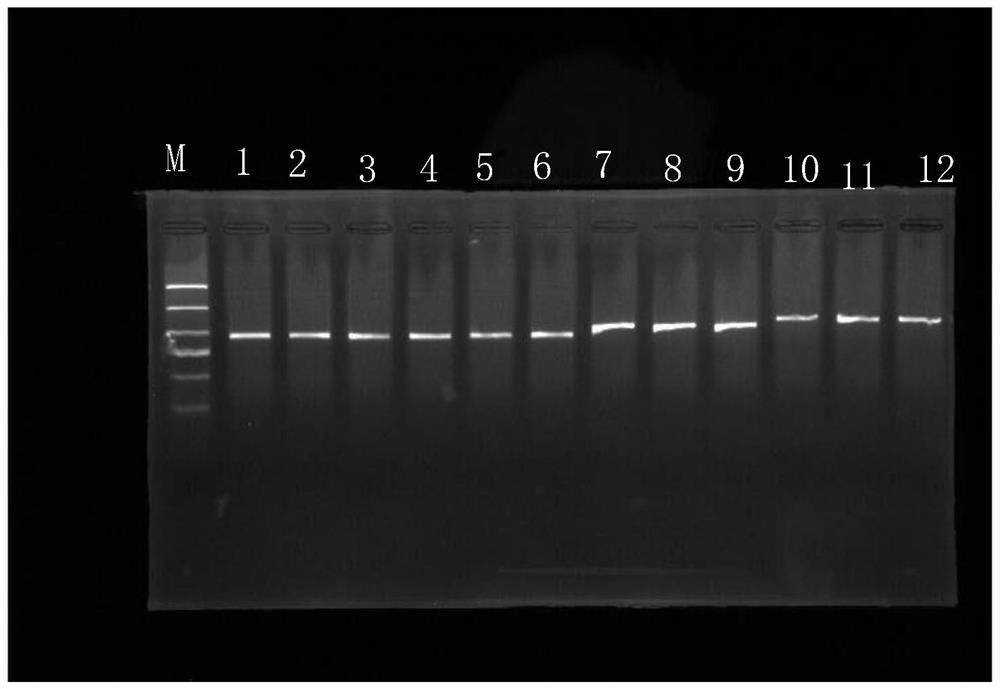
Image
Examples
Embodiment Construction
[0024] The technical solutions in the embodiments of the present invention will be clearly and completely described below in conjunction with the embodiments of the present invention and the accompanying drawings. Apparently, the described embodiments are only some of the embodiments of the present invention, not all of them. Based on the embodiments of the present invention, all other embodiments obtained by persons of ordinary skill in the art without making creative efforts belong to the protection scope of the present invention.
[0025] (1) Collection, identification and preservation of fish samples of Pleuriformes Soleidae
[0026] All the fish samples of Pleuriformes Soleidae used in the examples were collected from the natural environment in the wild, and all the collected specimens were brought back to the laboratory for information registration, and all were identified with a stereo microscope to determine the species. The samples of Plaiceiformes soles that had been...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com