A high-density molecular marker-assisted aggregation breeding method based on core pedigree varieties
A technology of molecular marker-assisted and aggregated breeding, which is applied in botany equipment and methods, biochemical equipment and methods, plant products, etc., can solve the problems of long-term, high investment, wide-spreading hybrid breeding, etc., to reduce breeding costs , Convenience of aggregation breeding, reduction of blindness and randomness
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0048] 1. Determine the genome segment information related to the traits of the core pedigree varieties.
[0049] Using the research method in section (4) of the background technology, more than 1,000 genome segments related to traits of the South China rice core pedigree variety "Huanghuazhan" were determined. In the subsequent analysis and processing, these genome segments are considered as the smallest recombination unit and are closely related to breeding.
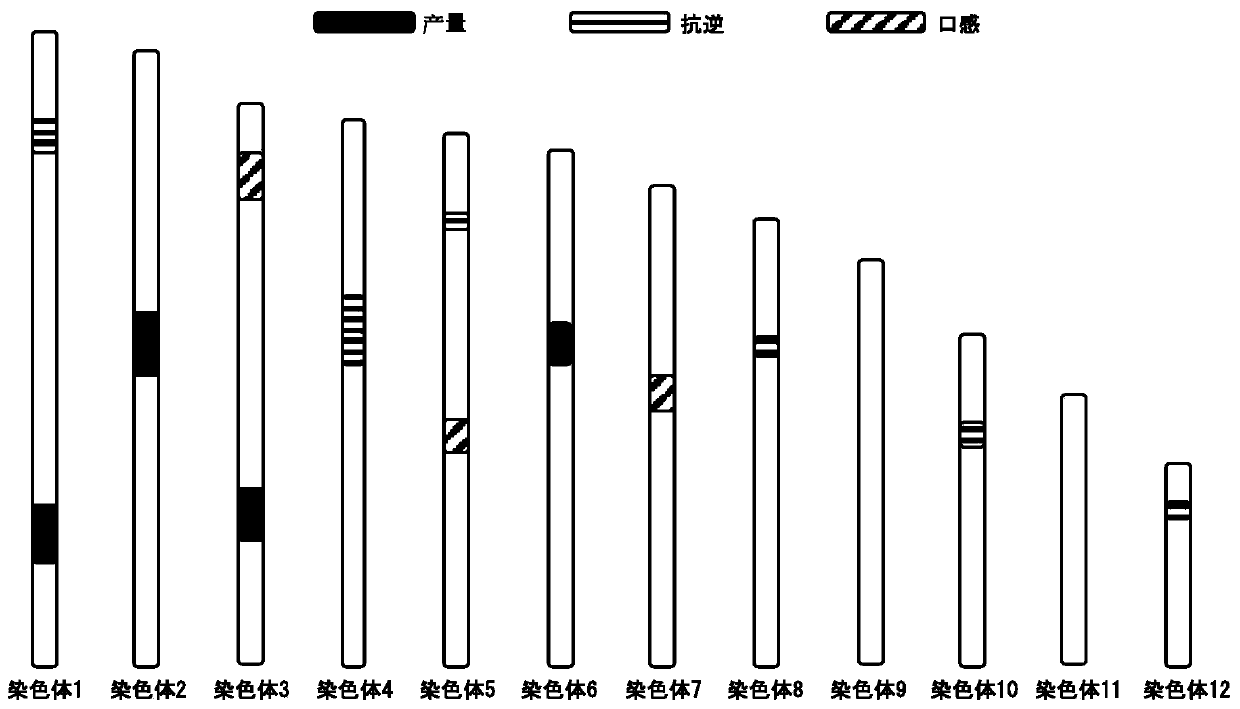
[0050] The literature [Pedigree-based analysis of derivation of genome segments of anelite rice reveals key regions during its breeding. Plant Biotechnology Journal, 2016, 14(2): 638-648] introduced in detail the method of population resequencing combined with recombination breakpoint map (bin map) analysis, QTL mapping, and cloned gene information to determine the research methods of the associated segments of "Huang Huazhan" traits. Examples of related results are as follows figure 1 shown.
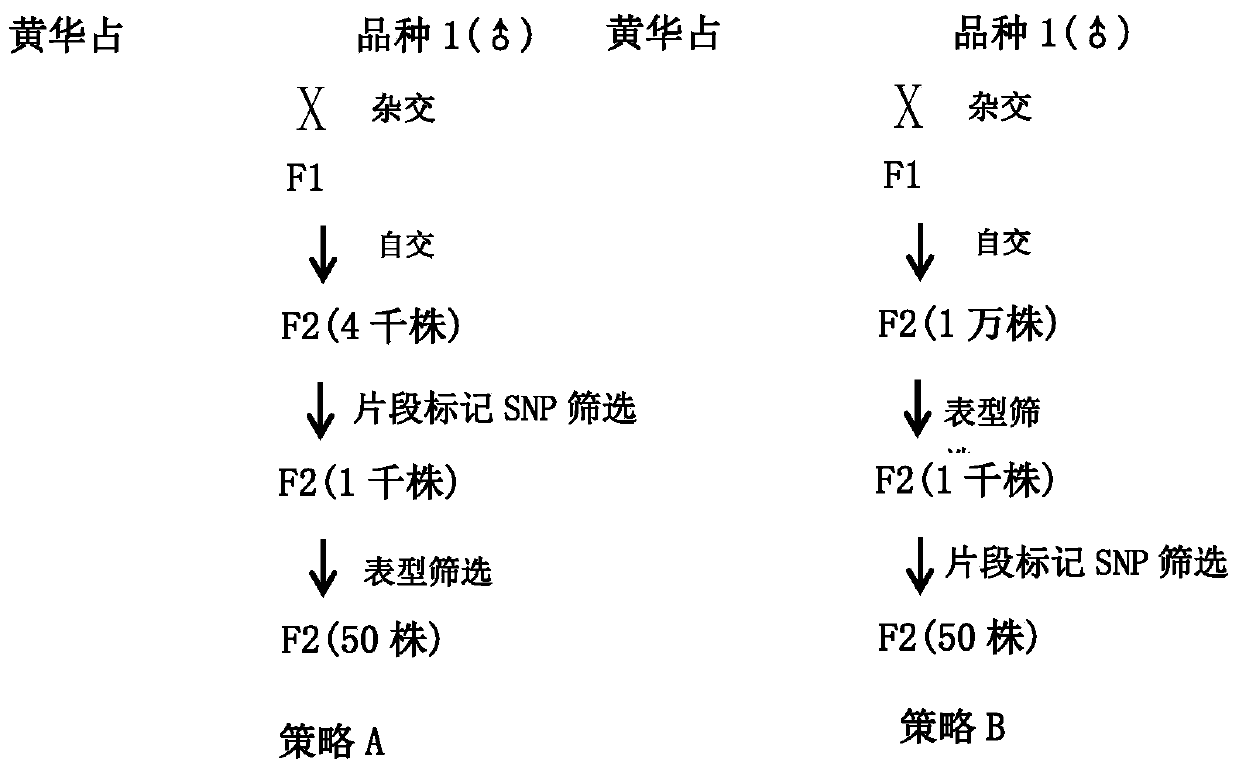
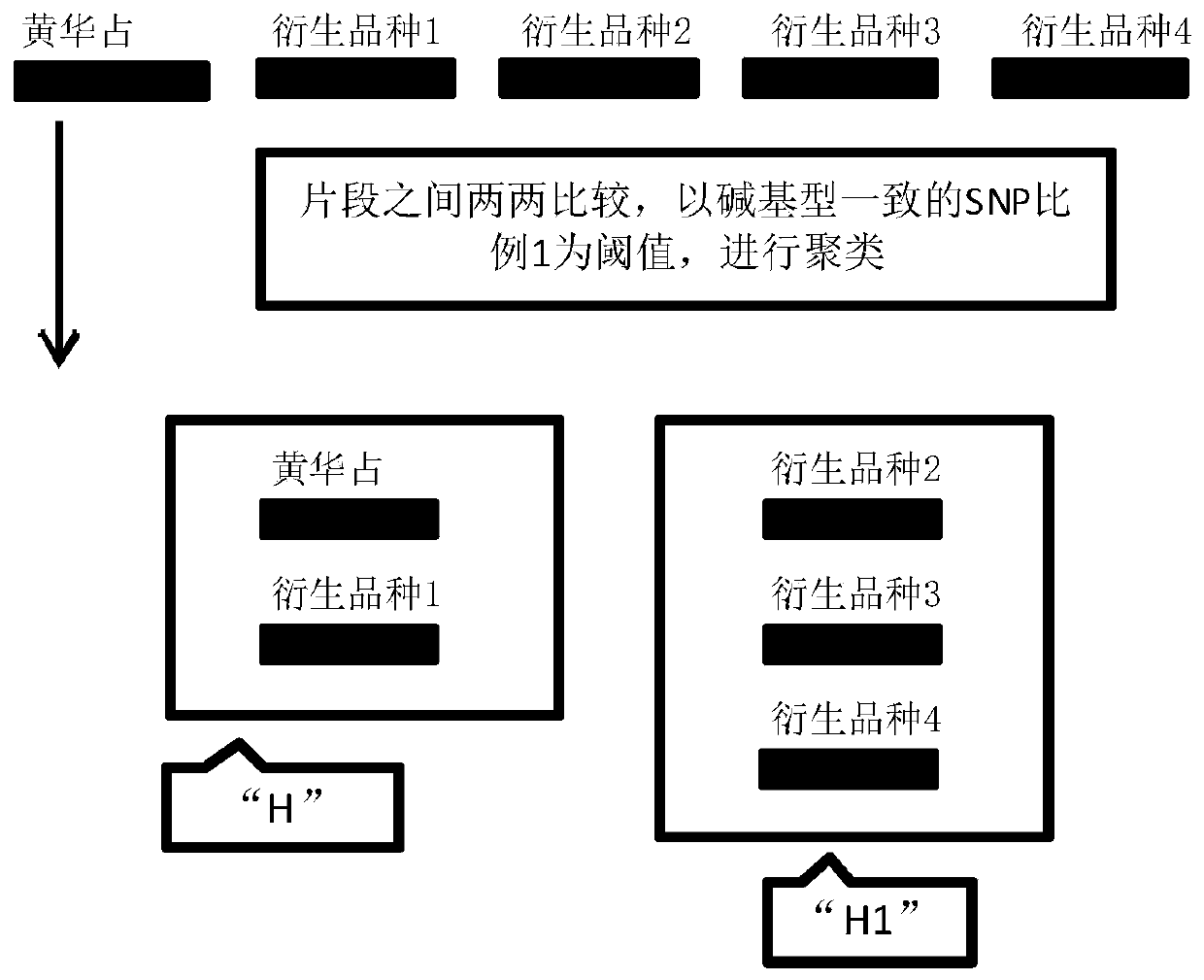
[0051] 2. Obtain the ...
Embodiment 2
[0098] In this example, the new-generation core pedigree variety Huangguangyouzhan derived from Huanghuazhan was used as the female parent, the introduced variety 1 was Wuguangzhan, and the introduced variety 2 was Fengyue Huazhan. Huangguangyouzhan is highly resistant to rice blast, the rice quality is not high-quality, and moderately sensitive to bacterial blight; Wuguangzhan is resistant to rice blast, with national standard high-quality grade 3 and provincial standard high-quality grade 2, and moderate resistance to bacterial blight; Fengyuehua Accounting: medium resistance to rice blast, rice quality national standard provincial standard high-quality grade 2, medium resistance to bacterial blight. The purpose of breeding is to maintain the high yield and rice blast resistance of Huangguangyouzhan, introduce the high-quality rice traits and bacterial blight resistance of Wuguangzhan and Fengyue Huazhan, and select high-yield, high-quality, and disease-resistant conventional...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com