A Mass Spectrometry Method for the Interaction of Active Proteins and Small Molecules
A mass spectrometry detection and protein technology, which is applied in the field of interaction detection between active proteins and small molecules, can solve the problems of long analysis cycle, difficult purification, time-consuming and laborious, etc.
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
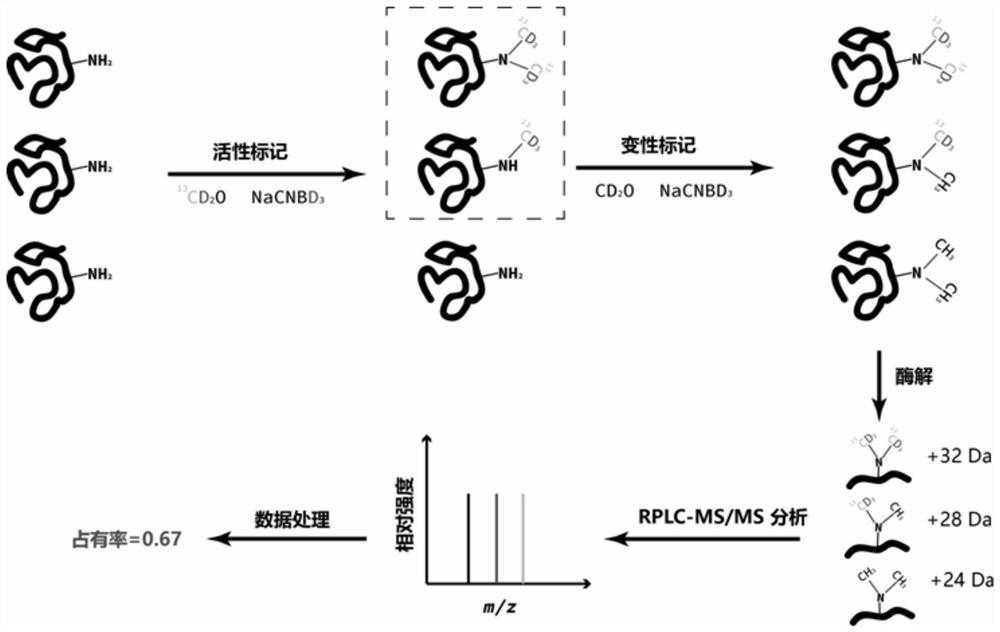
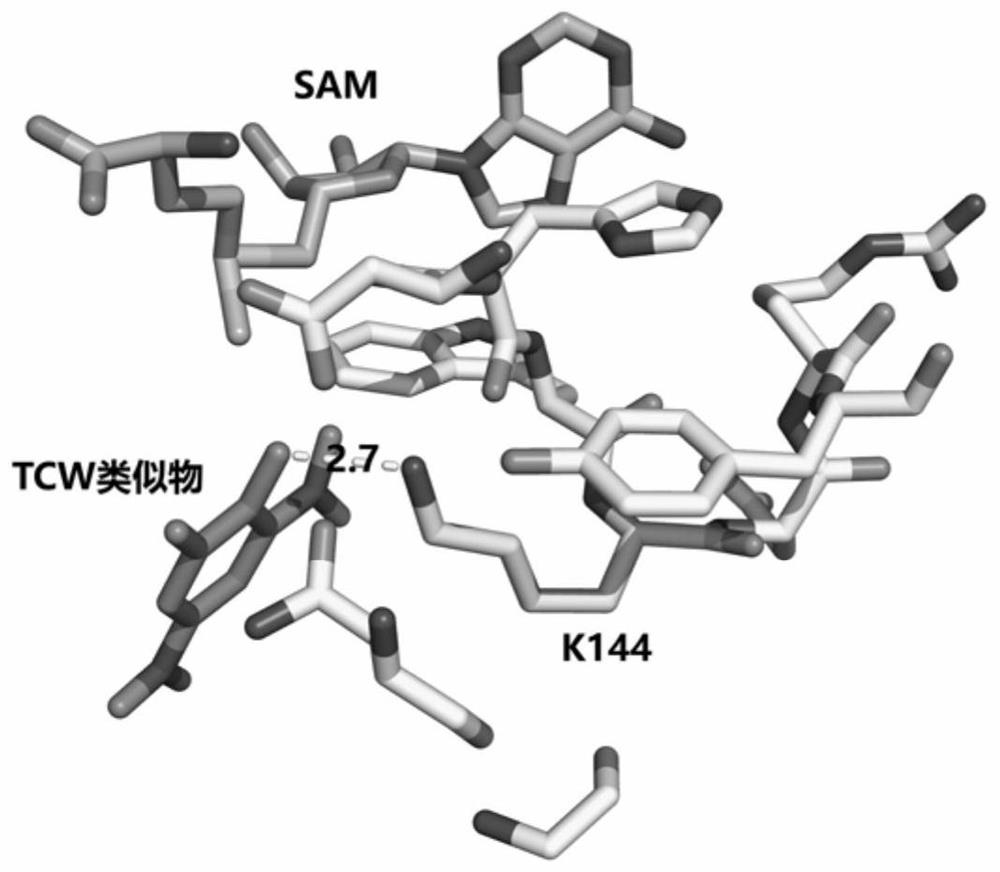
[0039] Detection of Interaction between Catechol-O-methyltransferase (COMT) and Small Molecule Tolcapone (TCW)
[0040] Each of the purified catechol methyltransferases was dissolved in the following three reaction systems to keep the protein concentration at 1 mg / mL. The reaction systems were: 1) 50 mM phosphate buffered saline (PBS buffer), 1.6 mM Thiothreitol (dithiothreitol, DTT), pH 7.4; 2) 50mM PBS buffer, 1.6mM DTT, pH 7.4, 5mM MgCl 2 and 200μM S-adenosyl methionine (SAM); 3) 50mM PBS buffer, 1.6mM DTT, pH 7.4, 5mM MgCl 2 and 200 μM S-adenosyl methionine (SAM) and 10 μM TCW, and incubated on a temperature-controlled shaker at 25 degrees for 30 min. Add 10 mM C to the above three samples 13 D. 2 O and 15 mM NaCNBD 3 Carry out the dimethylation labeling reaction in the active state, the temperature is room temperature, and the reaction time is controlled at 30 minutes. After the reaction, 5 times the volume of protein precipitation solution and 50 mM ammonium acetate ...
Embodiment 2
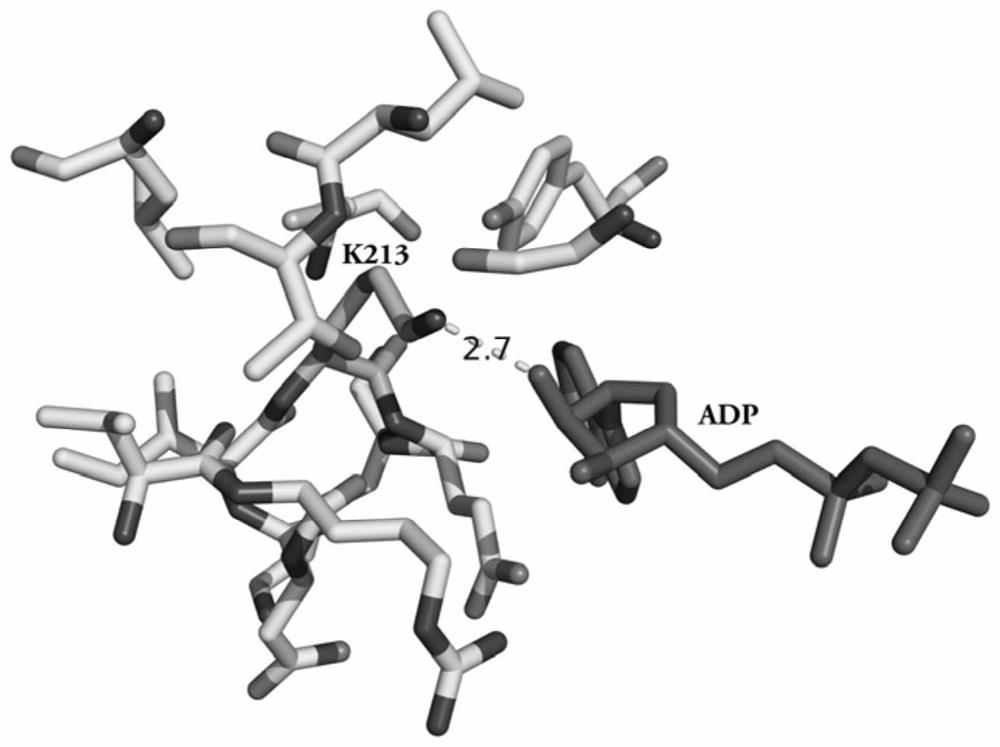
[0044] Analysis of ATP-binding proteins in complex biological samples
[0045] Take 10,000,000 cells and disperse in 1mL active lysate, the active lysate components are 50mM Hepe buffer, 1mM EDTA, 150mM NaCl, 5% Glycerol, 1% NP-40, 1mM phenylmethylsulfonyl fluoride (PMSF). Under the condition of ice bath, ultrasonic-assisted cell disruption was performed, the ultrasonic power was 200W, the ultrasonic program was set to ultrasonic time 3S, interval time 5S, ultrasonic 30 times. Ultrasonic disrupted cells were subjected to high-speed centrifugation at 4°C to remove cell debris and other insoluble matter with a centrifugal force of 10,000 g and a centrifugation time of 10 min. The BCA protein concentration assay was used to determine the extracted protein concentration, and 6 parts of 200 μg extracted protein were taken respectively and divided into 2 groups (experimental group and control group), with 3 samples in each group. Dilute the protein concentration of each sample to ...
Embodiment 3
[0048] Interaction detection between bovine serum albumin (BSA) and 8-anilino-1-naphthalene sulfonate (ANS)
[0049] Each BSA was dissolved in the following three reaction systems to keep its protein concentration at 10mg / mL. The reaction systems were: 1) 100mM phosphate buffer (PBS buffer), 0.15mM ANS, pH 6.8; 2) 100mM PBS buffer , 0.6mM ANS, pH 6.8; 3) 100mM PBS buffer, pH 6.8, incubated on a 37-degree temperature-controlled shaker for 60min. Add 50mM C to the above three samples 13 D. 2 O and 50 mM NaCNBD 3Carry out the dimethylation labeling reaction in the active state, the temperature is 37 degrees, and the reaction time is controlled at 15 minutes. After the reaction, 4 times the volume of protein precipitation solution and 50 mM ammonium acetate were added, and the BSA protein was precipitated in a -20 degree refrigerator for 10 h. The precipitated protein was separated by high-speed centrifugation at 25000 g for 30 min at 4 degrees, and the precipitated protein was...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com