Method for achieving gene knockout by utilizing base editing technique and application thereof
A gene knockout and base editing technology, applied in chemical instruments and methods, recombinant DNA technology, biochemical equipment and methods, etc., can solve the problems that the effect of primary cells and hypermethylation sites needs to be improved
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
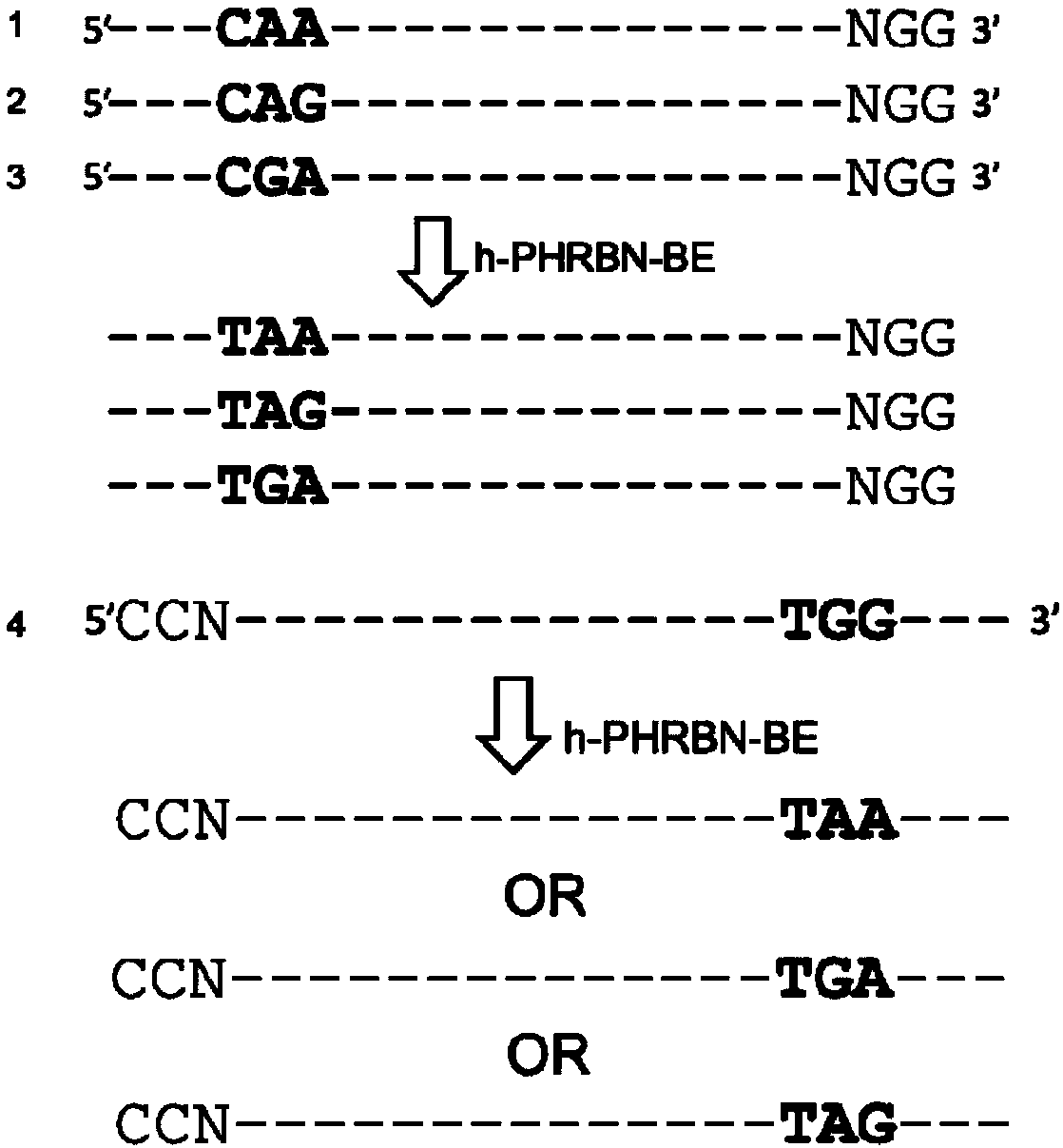
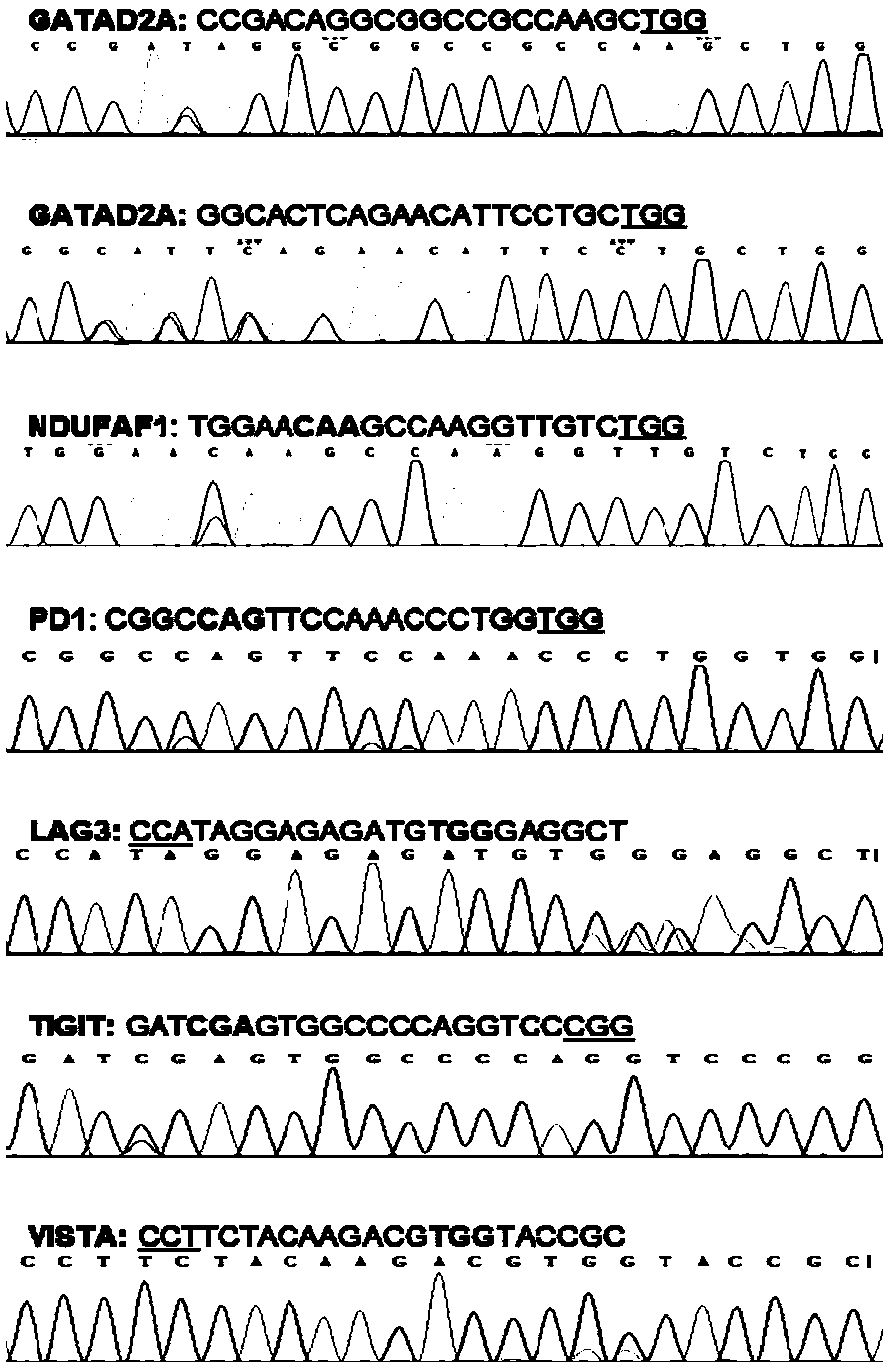
[0071] Base editing mediated by h-PHRBN-BE was performed on human cell lines, and stop codons were introduced to achieve gene knockout. Carry out gene knockout of cell lines (by electroporation or liposome transfection) according to routine operation, taking liposome transfection as an example. This embodiment selects SEQ ID NO.4-SEQ ID NO.10 as an example for illustration.
[0072] 1.1. Plasmid construction
[0073] For the selected 6 human-related gene sgRNAs, take the 20bp sequence in front of the PAM sequence and synthesize oligo, in which the upstream sequence is added with 5'-accg-3', the reverse complementary sequence is added with 5'-aaac-3', and the upstream and downstream sequences are passed Program (95°C, 5min; 95°C-85°C at-2°C / s; 85°C-25°C at-0.1°C / s; hold at 4°C) annealing, connected to the BsaI (NEB:R0539L) linearized pGL3-U6sgRNA (addgene: 51133) vector. The linearization system is as follows: pGL3-U6sgRNA 2μg; buffer (NEB:R0539L) 6μL; BsaI 2μL; ddH 2 O to ...
Embodiment 2
[0081] The current problem of the BE3 system is that the efficiency is low at highly methylated sites, while h-PHRBN-BE still exhibits high activity at highly methylated sites, so we chose the highly methylated FANCF site to design three sgRNAs (SEQ ID NO.11-SEQ ID NO.13), used to compare the gene knockout efficiency of BE3 and h-PHRBN-BE. Carry out gene knockout of cell lines (by electroporation or liposome transfection) according to routine operation, taking liposome transfection as an example.
[0082] 1.1. Plasmid construction
[0083]For the selected 6 human-related gene sgRNAs, take the 20bp sequence in front of the PAM sequence and synthesize oligo, in which the upstream sequence is added with 5'-accg-3', the reverse complementary sequence is added with 5'-aaac-3', and the upstream and downstream sequences are passed Program (95°C, 5min; 95°C-85°C at-2°C / s; 85°C-25°C at-0.1°C / s; hold at 4°C) annealing, connected to the BsaI (NEB:R0539L) linearized pGL3-U6sgRNA (addgen...
Embodiment 3
[0090] Another problem currently existing in the BE3 system is that the editing efficiency of the target site C is affected by the previous G, that is, if the G is in front of the C, the editing efficiency will be greatly reduced, but the h-PHRBN-BE system does not have such a problem, so we chose For two genes, a total of four sgRNAs (SEQ ID NO.14-SEQ ID NO.17) were designed to compare the knockout efficiency of BE3 and h-PHRBN-BE. Carry out gene knockout of cell lines (by electroporation or liposome transfection) according to routine operation, taking liposome transfection as an example.
[0091] 1.1. Plasmid construction
[0092] For the selected 6 human-related gene sgRNAs, take the 20bp sequence in front of the PAM sequence and synthesize oligo, in which the upstream sequence is added with 5'-accg-3', the reverse complementary sequence is added with 5'-aaac-3', and the upstream and downstream sequences are passed Program (95°C, 5min; 95°C-85°C at-2°C / s; 85°C-25°C at-0.1°...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com