A kind of petroleum degrading bacterium capable of degrading heavy crude oil and its separation method and application
A petroleum-degrading bacteria and petroleum-degrading technology, applied in the field of petroleum-degrading bacteria and its separation, can solve the problems of less attention to degradation of crude oil, limited utilization of petroleum hydrocarbon substrates, and difficult degradation, etc., achieving low application cost and high degradation efficiency , The effect of low process requirements
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0040] Domestication, isolation and purification methods of petroleum degrading bacteria:
[0041]1) Put 5g of petroleum-contaminated soil samples into a 50mL centrifuge tube, add 25mL of ultrapure water, and shake on a vortex shaker for 5-10min to fully disperse the soil and let it settle. Among them, the oil-contaminated soil samples came from an oil-contaminated site.
[0042] 2) Take 10 mL of the supernatant and add it to 100 mL of petroleum liquid medium, and culture it on a shaker at 30° C. and 150 r / min for 7 days.
[0043] 3) After 7 days, inoculate at 5%, and re-transfer into fresh petroleum liquid medium. The culture conditions are the same as above, and the enrichment culture is continuously transferred for 3 times.
[0044] 4) Separation is carried out by the dilution coating plate method, and the culture solution is diluted to 10 -3 or 10 -4 Afterwards, 100 μL of the diluted bacterial solution was applied to fresh solid isolation medium.
[0045] 5) Cultivate ...
Embodiment 2
[0049] Molecular biological identification of strains
[0050] 1) Streak culture the preserved strain in LB solid medium, extract the whole genome with a DNA kit after culture, and perform PCR amplification with primers 27-F and 1492-R. The PCR reaction system was 25 μL: 1 μL of upstream and downstream primers (10 μmol / L), 1 μL of DNA template (10 ng / μL), 12.5 μL of 2×Taq Master Mix, and made up to 25 μL with ultrapure water. The PCR reaction conditions were: 94°C for 5 min; 94°C for 1 min, 52°C for 1 min, 72°C for 2 min, 30 cycles; 72°C for 10 min.
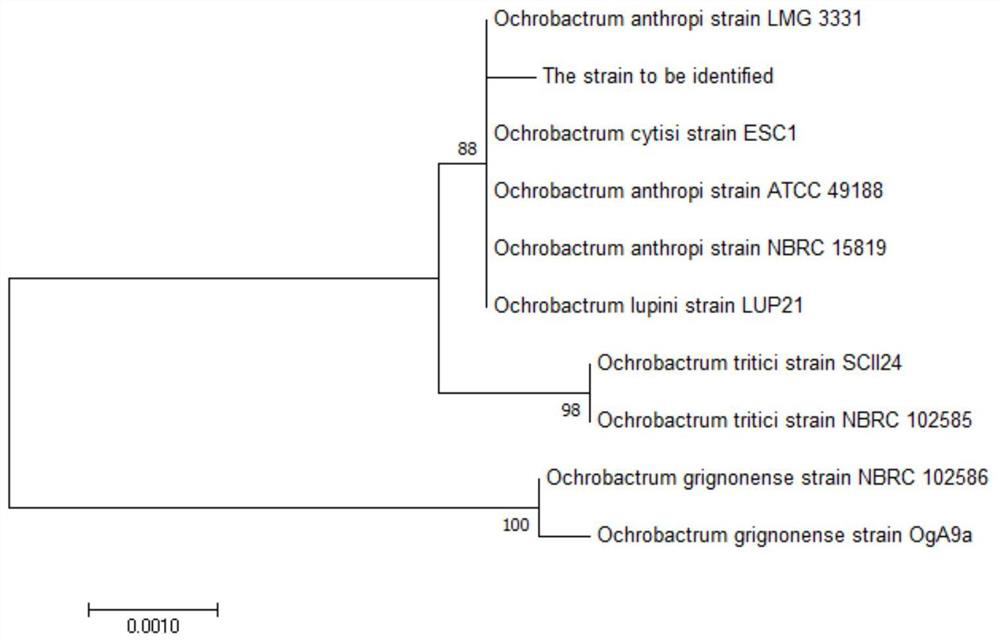
[0051] 2) The PCR amplification product was entrusted to Sangon Bioengineering (Shanghai) Co., Ltd. for sequencing, and the obtained gene sequence was compared with GenBank to determine its taxonomic status.
[0052] The 16S rRNA gene sequencing result of the strain is shown in SEQ ID NO: 1, the result shows that the gene sequence length is about 1348 (sequence), and the sequence comparison results show that it is similar to the...
Embodiment 3
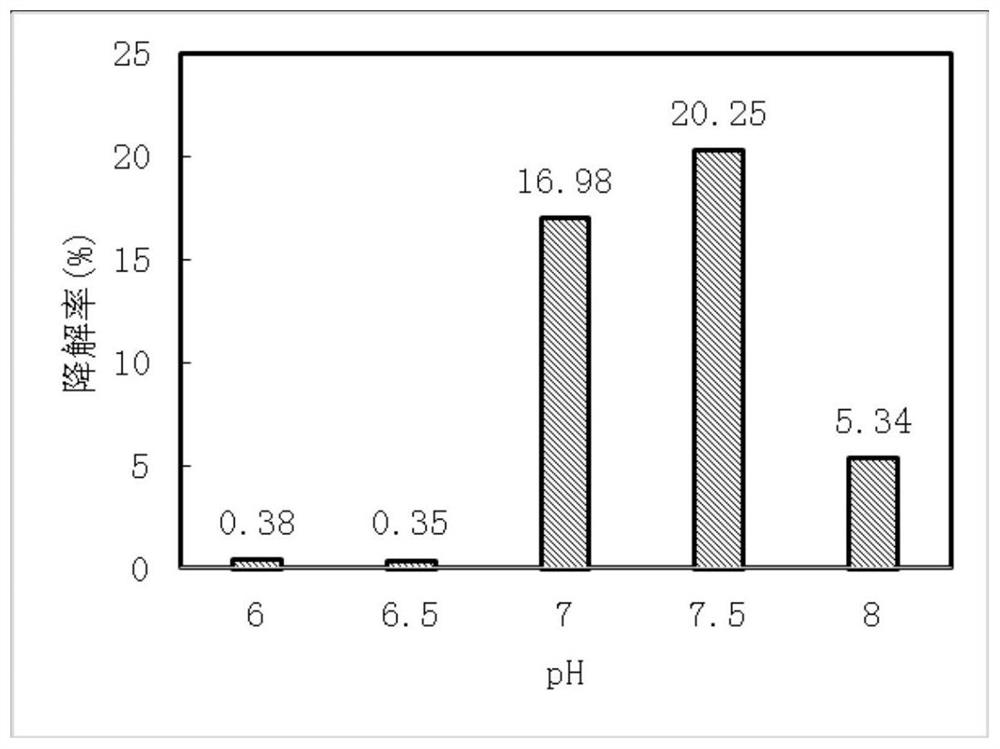
[0056] Method for Determination of Petroleum Degradation Rate
[0057] 1) Add 0, 0.5, 1, 2, 3, 4mL standard oil solution to 5 volumetric flasks respectively, and dilute to the mark line with petroleum ether.
[0058] Pour the prepared standard solution into a quartz cuvette, use petroleum ether as a reference, and use a UV spectrophotometer to measure the absorbance at a wavelength of 226nm. Take the absorbance as the ordinate, and the concentration of crude oil as the abscissa, and draw after blank correction standard curve line.
[0059] 2) Inoculate the preserved strains back into 100mL petroleum liquid medium, and culture them on a shaking table at 30°C and 150r / min for 7 days (set up a sterile control group at the same time), extract undegraded petroleum in the medium with petroleum ether, and use ultraviolet spectroscopy to Measure the absorbance with a photometer at a wavelength of 226nm, and calculate the remaining oil concentration with reference to the standard curv...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com