Methods for assessing risk of developing colorectal cancer
A colorectal cancer, risk technology, applied in the direction of biochemical equipment and methods, microbial measurement/testing, instruments, etc., can solve the problem of complex genetic susceptibility to colorectal cancer
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
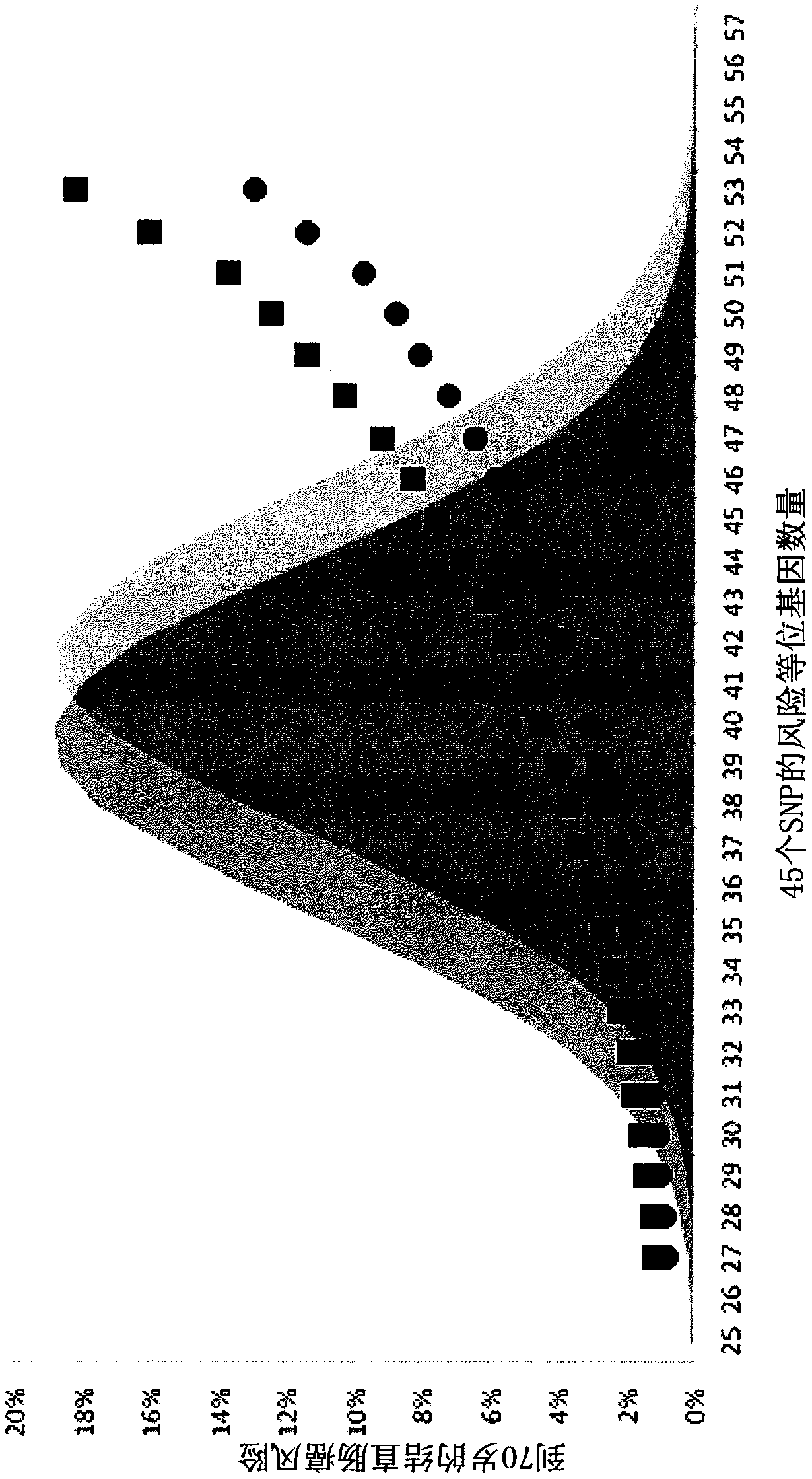
[0222] Example 1 - SNPs indicative of colorectal cancer risk
[0223] Fifty-four SNPs associated with colorectal cancer in a European population were identified. Among them, 4 SNPs within 11q12.2 (rs174537, rs4246215, rs174550, and rs1535) are fully related and can be represented by common haplotypes (herein referred to as 11q12.2 haplotypes). Two SNPs within 19q13.2 (rs1800469 and rs2241714) are perfectly related and can be represented by common haplotypes (referred to herein as 19q13.2 haplotypes). One SNP is located on the X chromosome (rs5934683) and was not included in colorectal cancer risk simulations for both males and females. Two SNPs (rs6687758 and rs6691170) within 1q41 are in linkage disequilibrium. Therefore, rs6691170 was excluded. Three SNPs (rs10505477, rs6983267 and rs7014346) within 8q24.21 had a D prime of 1.0. Therefore, rs10505477 and rs7014346 were excluded. The D prime of two SNPs (rs1035209 and rs11190164) within 10q24.2 was 0.9. Therefore, rs10...
Embodiment 2
[0230] Example 2 - Risk Allele Simulation
[0231] Simulations were performed using the software PLINK (Purcell et al., 2007) (http: / / pngu.mgh.harvard.edu / purcell / plink / ) to determine the cumulative number of risk alleles of SNPs that differentiate colorectal cancer cases from controls ability, and to estimate colorectal cancer risk as a function of the number of risk alleles.
[0232] A population of 1,000,000 people with colorectal cancer (cases) and 1,000,000 people without colorectal cancer (controls) was simulated. The distribution of SNP risk alleles for the simulated population matched the reported risk allele frequencies and per-allelic odds ratios for the colorectal cancer association. A simple risk model was assumed in this assessment in which the association of each SNP with colorectal cancer was independent. In this analysis, it was also assumed that the odds ratios for colorectal cancer reported by each SNP apply to both males and females and vary with age.
...
Embodiment 3
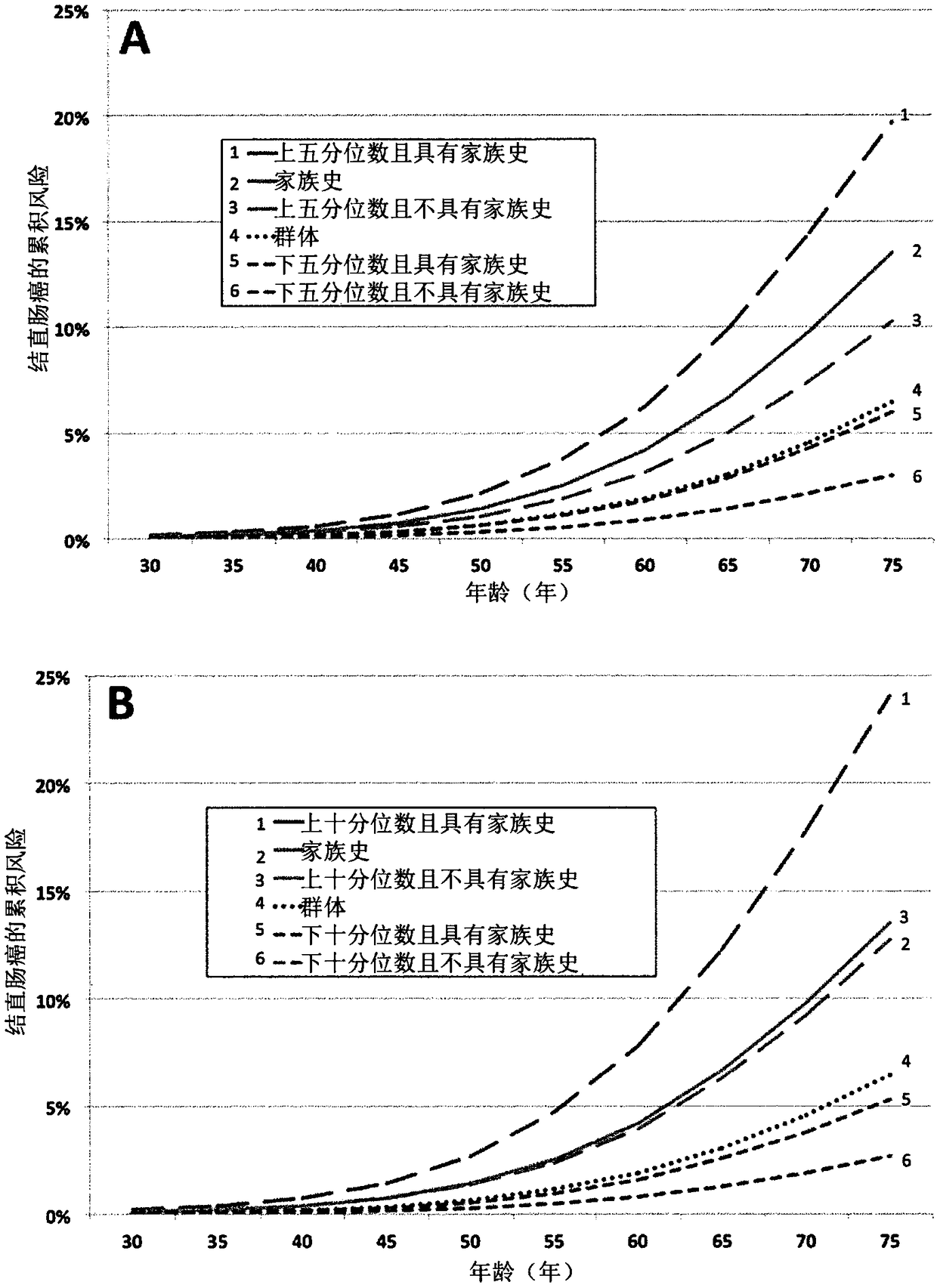
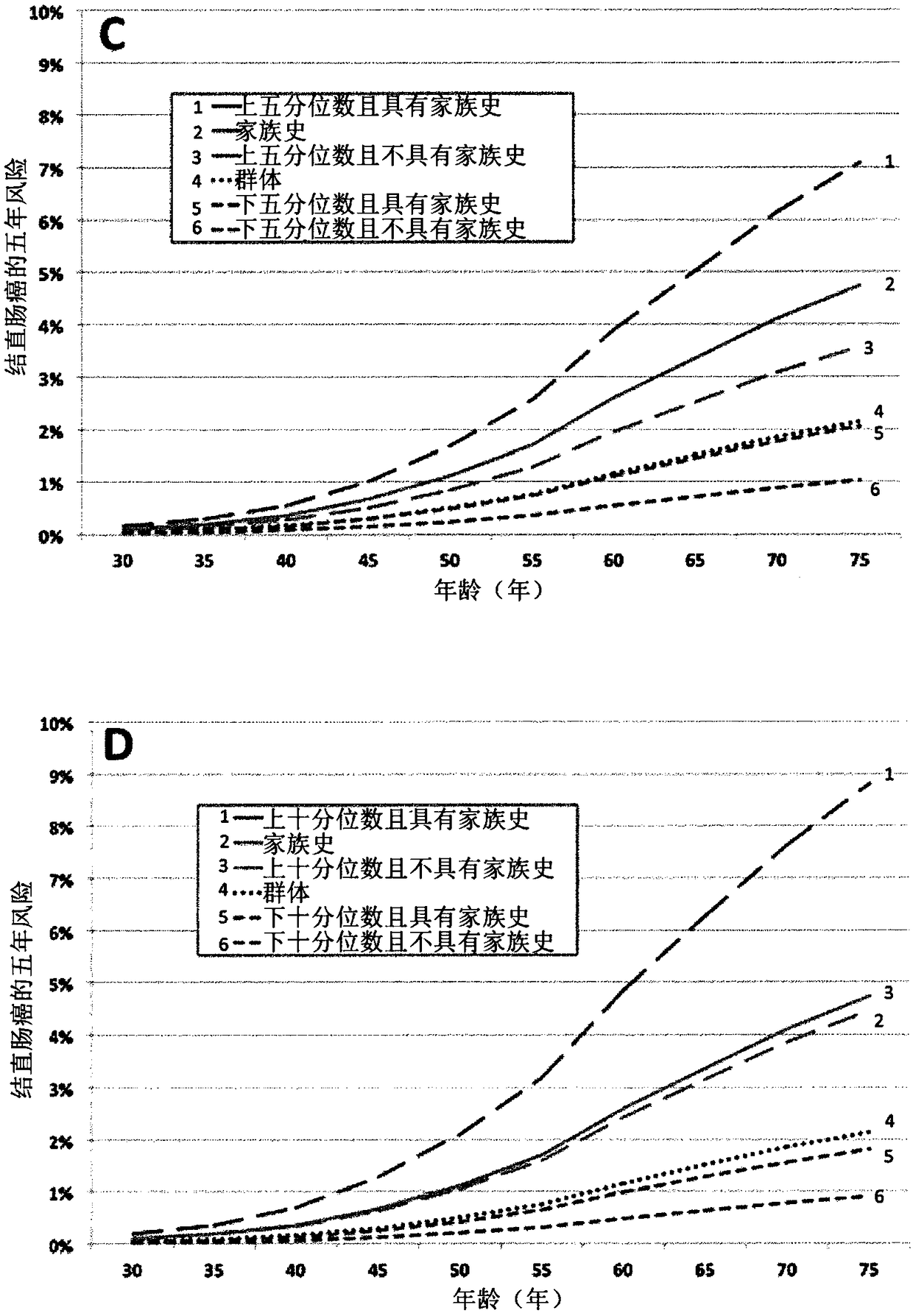
[0248] Example 3 - Classification of subjects according to risk of colorectal cancer
[0249] Simulations were used to quantify the utility of a panel of 45 risk-associated SNPs to classify persons according to their colorectal cancer risk. People at the ends of the risk allele spectrum were more likely to develop colorectal cancer (high end) or less likely to develop colorectal cancer (low end). Because the total variation in risk associated with these SNPs across the population can explain about a quarter of the total FRR, the predictive strength of the SNP profile increases if family history of colorectal cancer is also considered. Considering that for the bottom 20% of the population (for the number of risk alleles for these SNPs), the strength of the association with colorectal cancer was roughly inverse to the increased risk associated with the remaining FRR, with a family history of colorectal cancer, but at these Persons in the lowest quintile of the population for ...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com