Method for designing high-sensitivity and high-specificity mismatch nucleic acid sequences
A nucleic acid sequence and design method technology, applied in biochemical equipment and methods, microbial measurement/testing, etc., can solve the problem of low sensitivity and specificity of highly homologous sequence amplification, non-expandable highly homologous sequences, and dependence on personal experience and other issues to achieve the effect of improving the sensitivity of the amplification
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
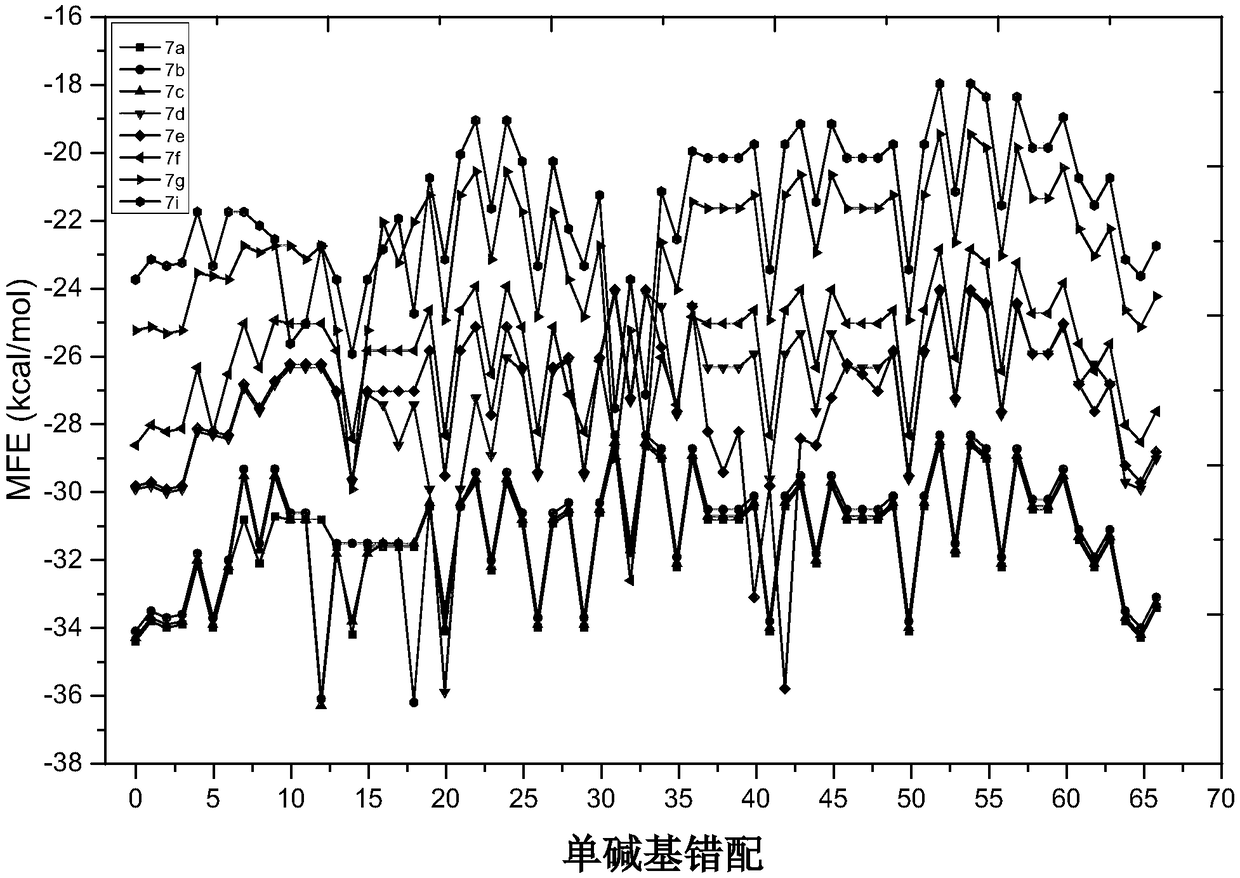
[0065] Example 1: Detection of let-7a
[0066] target sequence:
[0067] let-7a: 5'-UGAGGU AGU AGU AGG UUG UAU AGUU-3' (SEQ ID NO: 1)
[0068] Homologous Interfering Sequences:
[0069] let-7b:5'-UGAGGUAGUAGGUUGU G u G GUU-3' (SEQ ID NO: 2)
[0070] let-7c:5'-UGAGGUAGUAGGUUGUAU G GUU-3' (SEQ ID NO: 3)
[0071] let-7d:5'- A GAGGUAGUAGGUUG C AUAGU-3' (SEQ ID NO: 4)
[0072] let-7e:5'-UGAGGUAG G AGGUUGUAUAGU-3' (SEQ ID NO: 5)
[0073] let-7f:5'-UGA GGU AGU AG A UUG UAU AGU U-3 (SEQ ID NO: 6)
[0074] let-7g:5'-UGAGGUAGUAG U UUGUA C AGU-3' (SEQ ID NO: 7)
[0075] let-7i:5'-UGAGGU AGU AG U UUG UGC U GU U-3 (SEQ ID NO: 8)
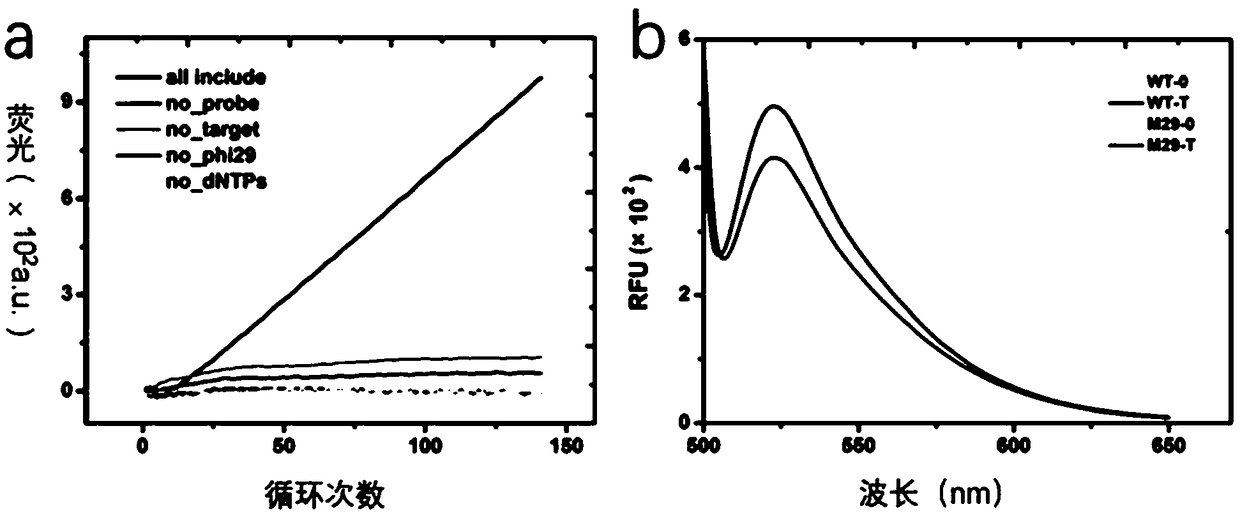
[0076] let-7a has 22 base sites, and each site of its probe chain has a combination of four bases: A, U, G, and C. For single base mismatches, the original base arrangement of let-7a is removed , there are 3×22=66 combinations in total. Using NUPACK software, based on the tendency difference of the secondary structure free energy of RNA, the...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com