Application of SpCas9n&PmCDA1&UGI base editing system to editing of plant genes
A technology for encoding genes and editing, applied in applications, plant peptides, plant products, etc., can solve problems such as limited scope, and achieve the effect of wide application prospects
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
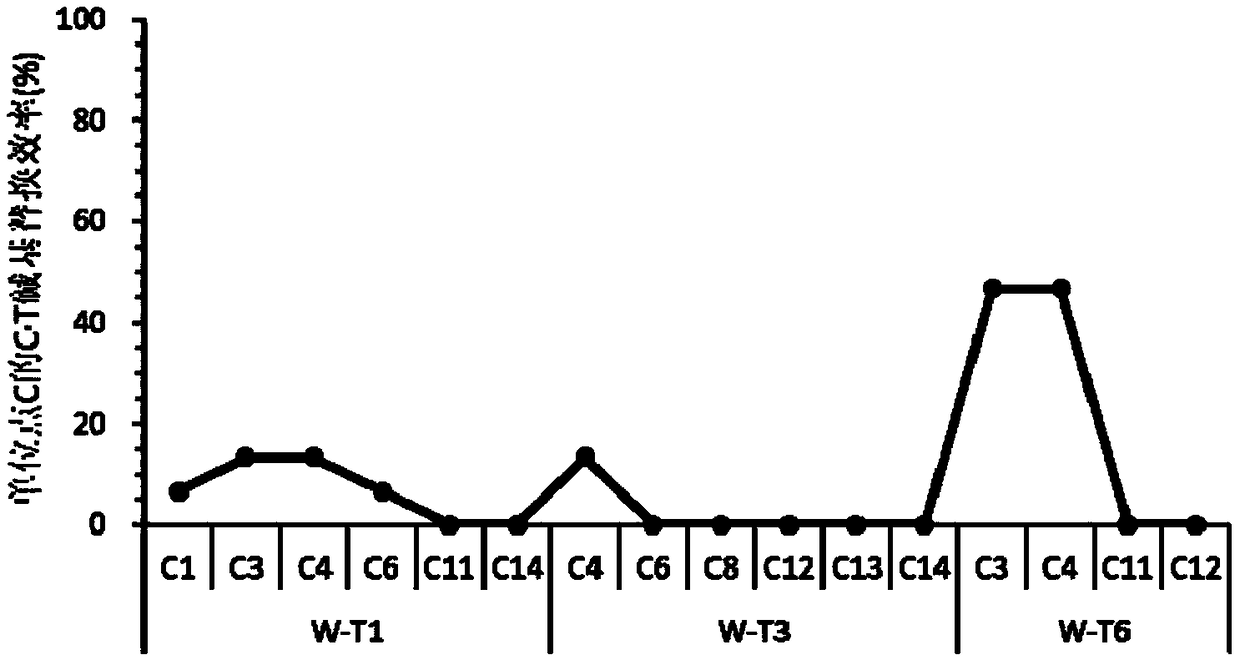
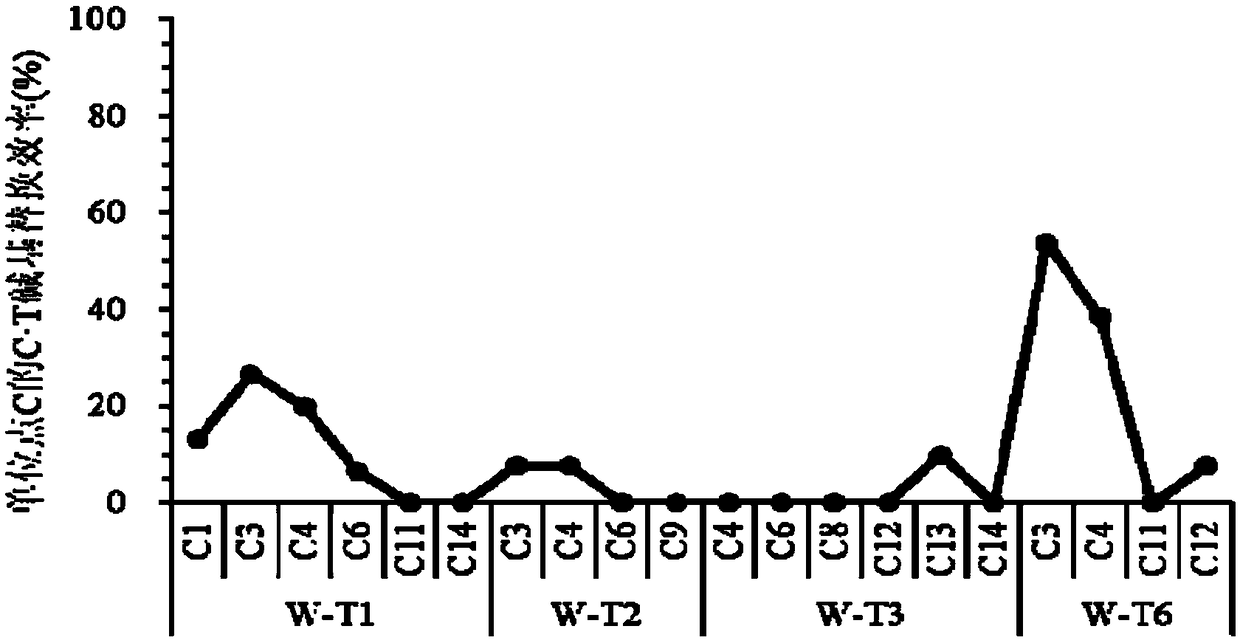
[0071] Example 1. The SpCas9n&PmCDA1&UGI base editing system can mutate C into T in the rice genome
[0072] 1. Construction of recombinant expression vector
[0073] Artificially synthesize the following recombinant vector SpCas9n&PmCDA1&UGI, which is a circular plasmid.
[0074] The sequence of SpCas9n&PmCDA1&UGI is sequence 1 in the sequence listing, the vector can express the protein SpCas9n shown in the sequence 2 in the sequence listing, the protein PmCDA1 shown in the sequence 3, the protein UGI shown in the sequence 4, and the six transcription targeting OsWaxy genes sgRNA (target points are marked as W-T1, W-T2, W-T3, W-T4, W-T5 and W-T6), the common sequence of these six sgRNAs is the sgRNA backbone, which is composed of the 571-646th sequence of sequence 1 bits are transcribed. Among them, the 1809-3522th position of sequence 1 is the OsUbq3 promoter sequence, the 3529-7797th position of sequence 1 is the coding sequence of SpCas9n, the 8089-8712th position is the...
PUM
 Login to view more
Login to view more Abstract
Description
Claims
Application Information
 Login to view more
Login to view more - R&D Engineer
- R&D Manager
- IP Professional
- Industry Leading Data Capabilities
- Powerful AI technology
- Patent DNA Extraction
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic.
© 2024 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap