Detecting sepsis
A sepsis and detection device technology, applied in the field of sepsis detection, can solve problems such as adverse drug reactions and increased antibiotic resistance
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0396] Example 1—Using CRP to diagnose sepsis
[0397] Although many new biomarkers for sepsis diagnosis have been investigated and are available in clinical practice, it is necessary to combine them with traditional markers, especially C-reactive protein (CRP), in order to increase sensitivity and specificity. This study compared the diagnostic accuracy of CRP alone and in combination with selected and complementary markers for the diagnosis of sepsis.
[0398] The method used:
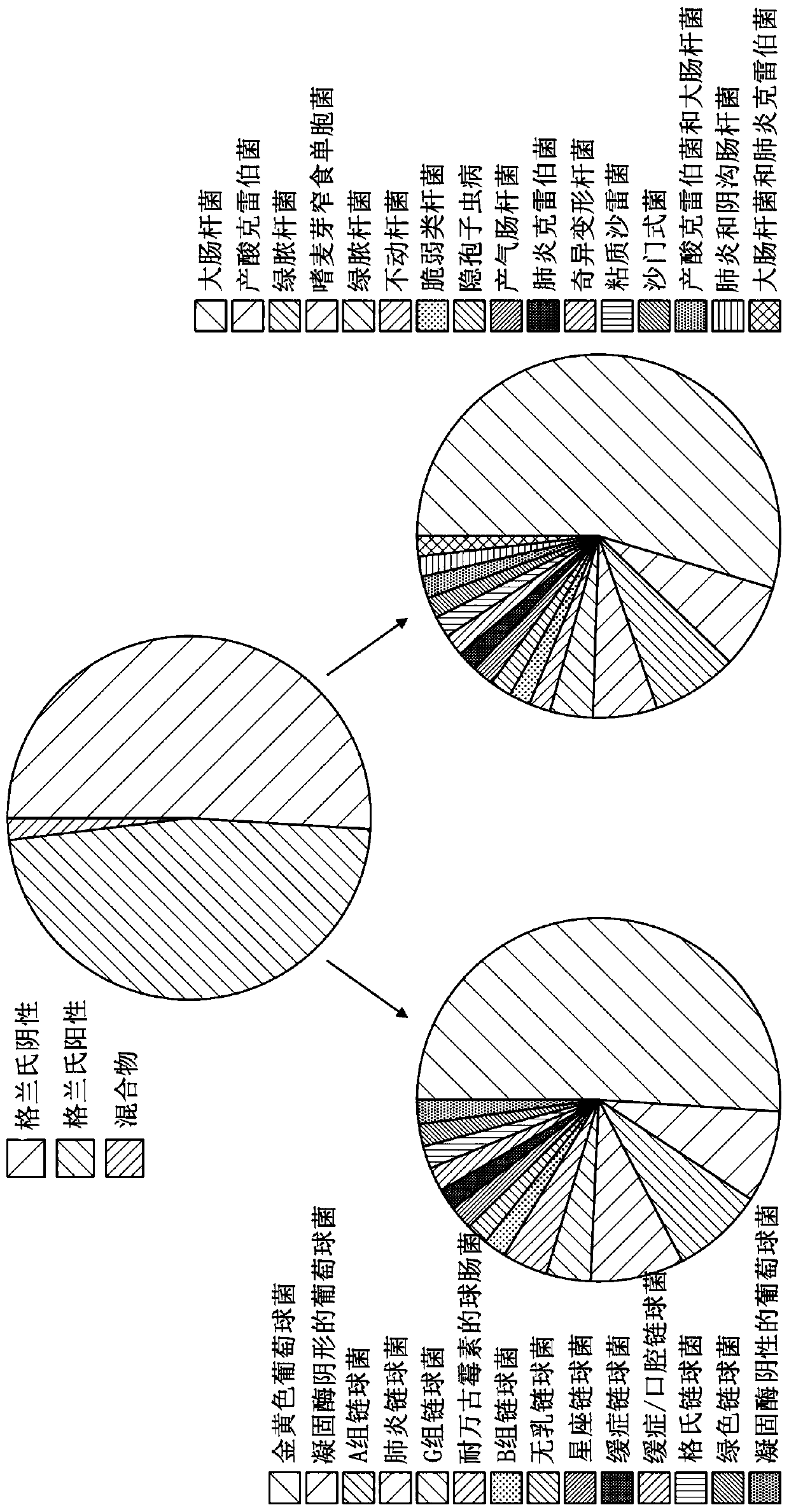
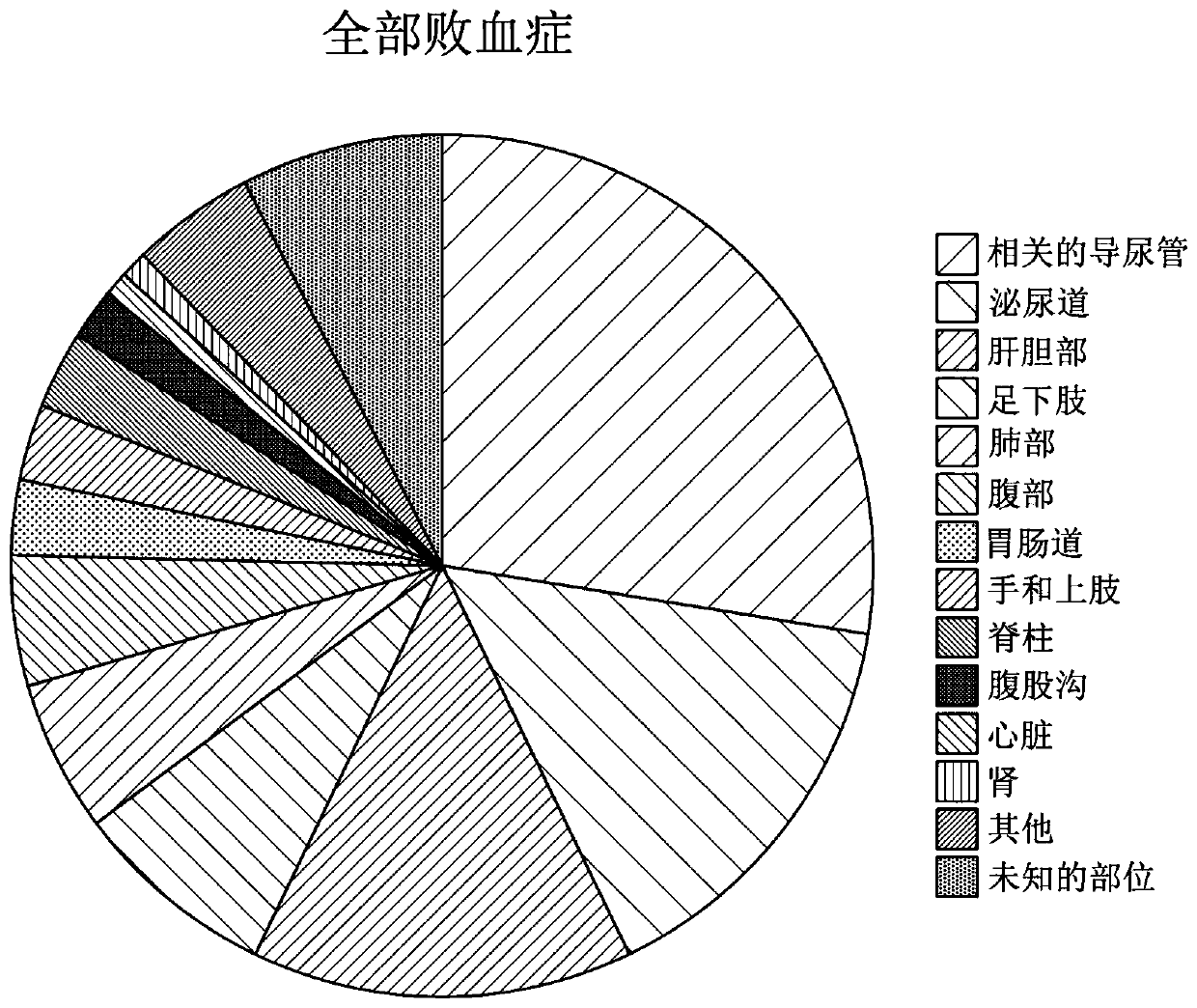
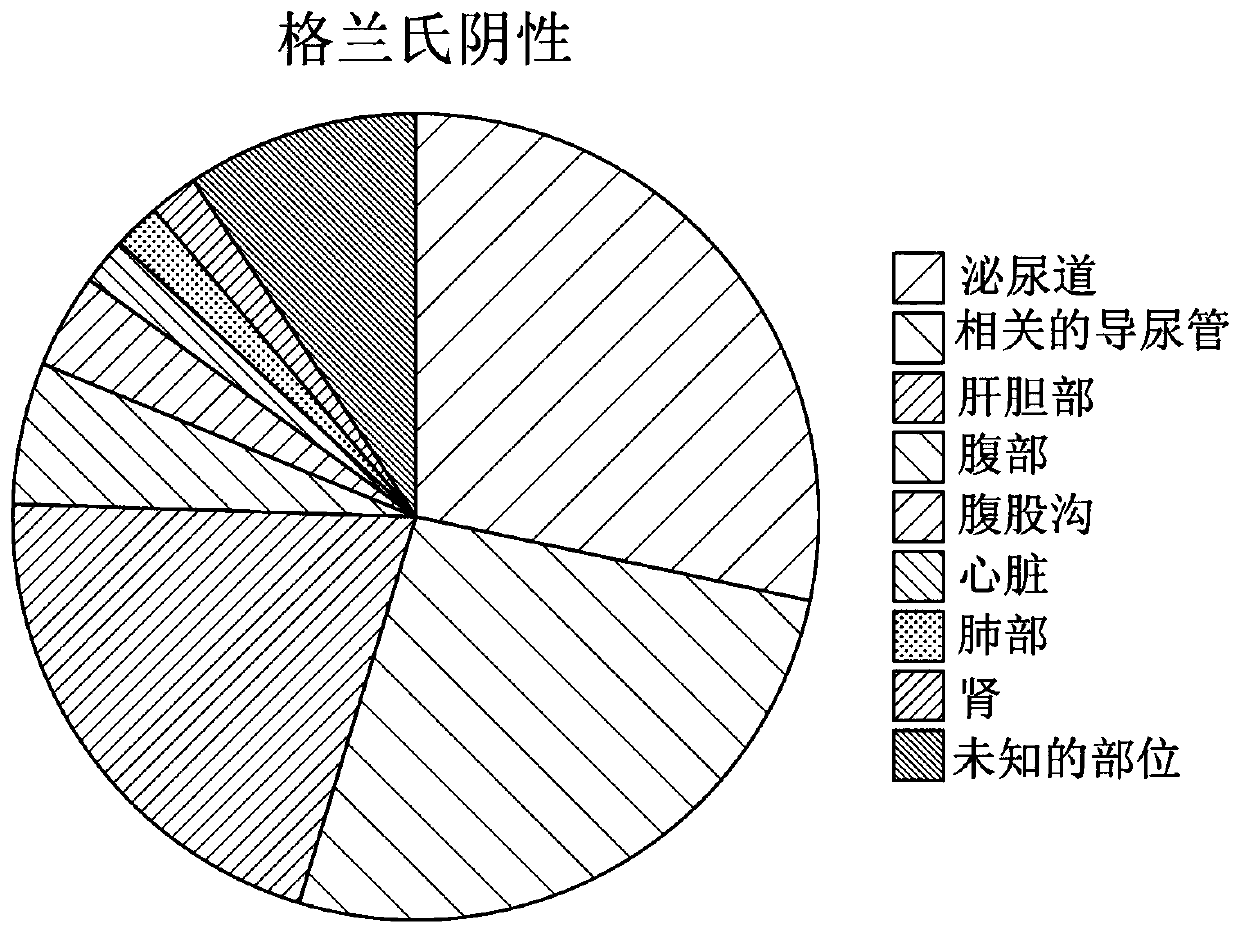
[0399] 102 patients with sepsis were diagnosed according to strict clinical criteria, including positive blood cultures (51 Gram-positive, 49 Gram-negative and 2 mixed Gram-positive and Gram-negative organisms) versus 102 without sepsis patients with evidence for comparison. Serum levels of CRP and procalcitonin (PCT), along with seven selected potential markers, were measured. These markers include: two inflammatory cytokines, interleukin-6 (IL-6) and tumor necrosis factor alpha (TNFα); the vas...
Embodiment 2
[0445] Example 2 - UHB Study and Results
[0446] Clinical samples were obtained from a clinical study conducted at the University of Birmingham Hospital and these samples were tested. Sort by sample time:
[0447] A = Registered admission / symptomatic onset
[0448] B = time required for blood culture (BC) *values are in bold (A / B sample number = 44)
[0449] C = BC positive time
[0450] D = Consent time (large volume) = 88 samples
[0451] E = last sample before release = 18 samples
[0452] Control = 102 samples
[0453] Use the following assay methods to detect:
[0454] Mark
Assay type
form
supplier
CRP (C-reactive protein)
ELISA
Sandwich
R&D Systems
CRP (C-reactive protein)
lateral flow
Sandwich
Mologic
sICAM1 (soluble intercellular adhesion molecule 1)
ELISA
Sandwich
R&D Systems
sICAM1 (soluble intercellular adhesion molecule 1)
lateral flow
Sandwich
Mologic
C...
Embodiment 3
[0468] Example 3 - DSTL Study and Results
[0469] 910 samples received were tested in 10 Mologic assays. Samples were collected from patients undergoing elective surgery and collected daily for up to 7 days after surgery. Patients were divided into 3 distinct groups:
[0470] 1. Control group (n=70) those recovered patients without SIRS symptoms
[0471] 2. SIRS group (n=66) those patients who developed SIRS symptoms in 7 days
[0472] 3. Sepsis group (n=70) those patients who developed sepsis in 7 days
[0473] Some samples were missing in cases where the patient refused consent or the study nurse was unable to obtain good venous access, for the sepsis group the focus was on days 1, 2 and 3 before sepsis.
[0474] Difference Between SIRS and Sepsis
[0475] Samples were grouped according to heart rate, respiratory rate, WCC and temperature, among other parameters, according to the SIRS diagnosis defined by DSTL. The first step was to analyze all samples from patients...
PUM
| Property | Measurement | Unit |
|---|---|---|
| Sensitivity | aaaaa | aaaaa |
| Sensitivity | aaaaa | aaaaa |
| Sensitivity | aaaaa | aaaaa |
Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com