Compositions and methods for characterizing solid tumors responsiveness to Anti-pd-l1 antibody monotherapy
A technology for PD-L1 and solid tumors, applied in chemical instruments and methods, antibodies, drug combinations, etc.
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
example 1
[0110] Example 1: CXCL9, CD274, LAG3, and IFNG are associated with objective response rate (ORR) in NSCLC
[0111] The safety and clinical activity of durvalumab in advanced solid tumors was evaluated. RNA sequencing data were generated on patient tumors with available clinical data, of which 30 had pre / post NSCLC patient tumor pairs and 30 bladder patient tumors were pretreated with available clinical data and subjected to bioinformatics analysis. The following genes were assessed: CXCL9, PD-L1(CD274), LAG-3, IFNG, PD-1(PDCD1), NKG7, CD8A, SLAMF7, PD-L2(PDCD1LG2), TIM-3(HAVCR2), CD80, CD86 , CTLA-4, CD2, TLR8, GZMK, TNFRSF4, FOXP3, CD276, B7-H4 (VTCN1 ), and CD37.
[0112] Of the 21 candidate genes evaluated, each gene was segmented into a low or high group using receiver operating characteristic (ROC) calculations using area under the curve (AUC), logistic regression, time-to-event analysis such as Kaplan- The calculations were performed with Mayer and Cox proportional haz...
example 2
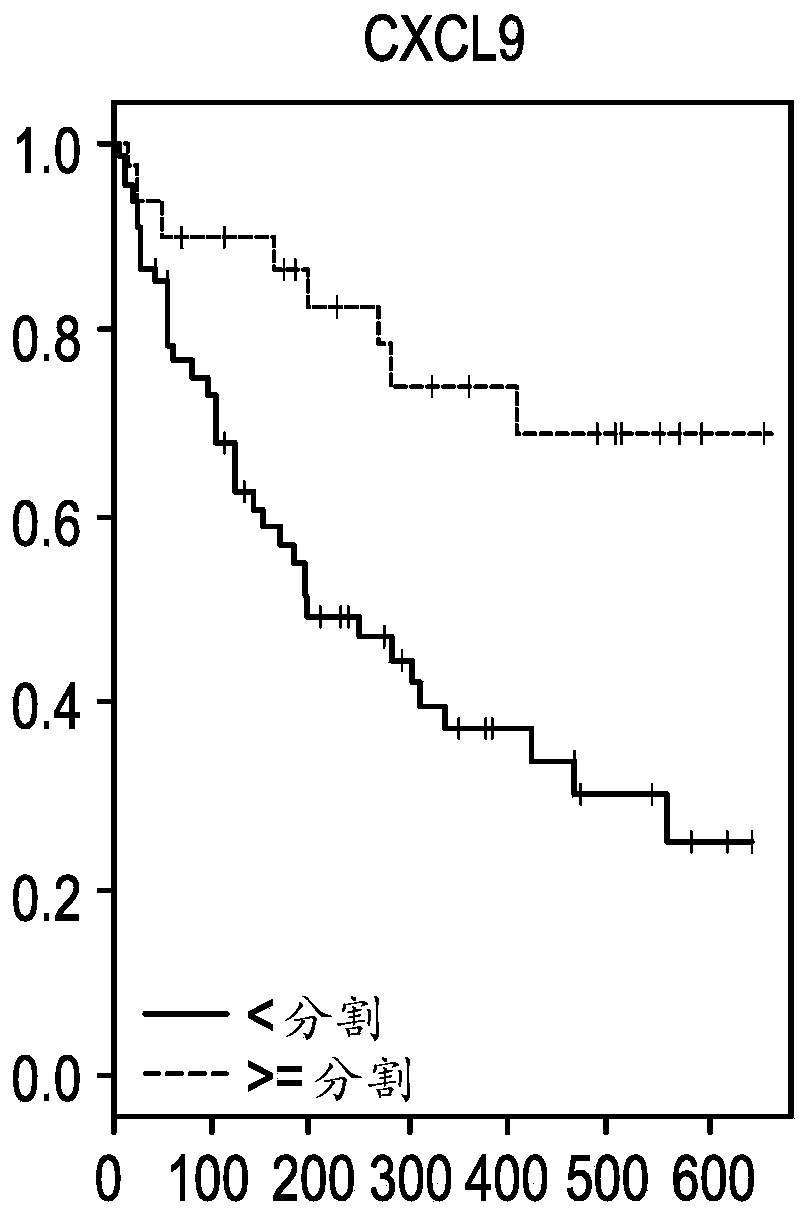
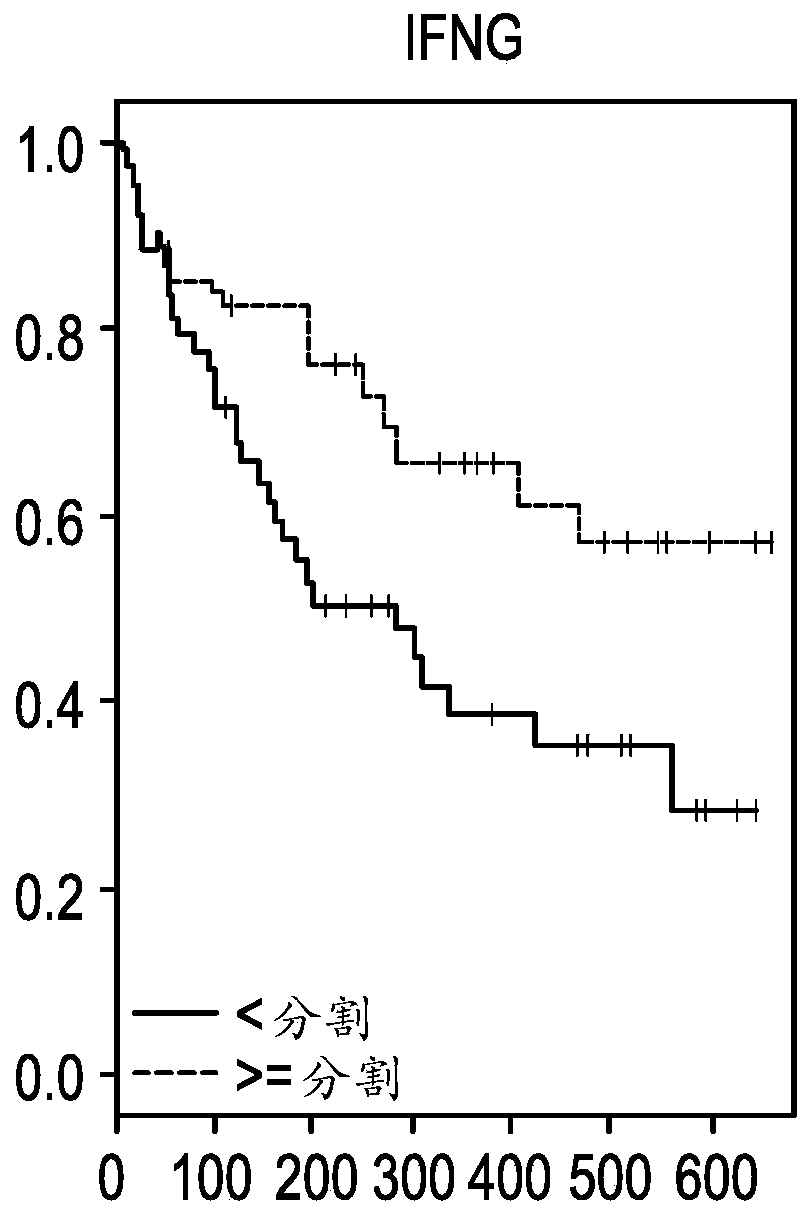
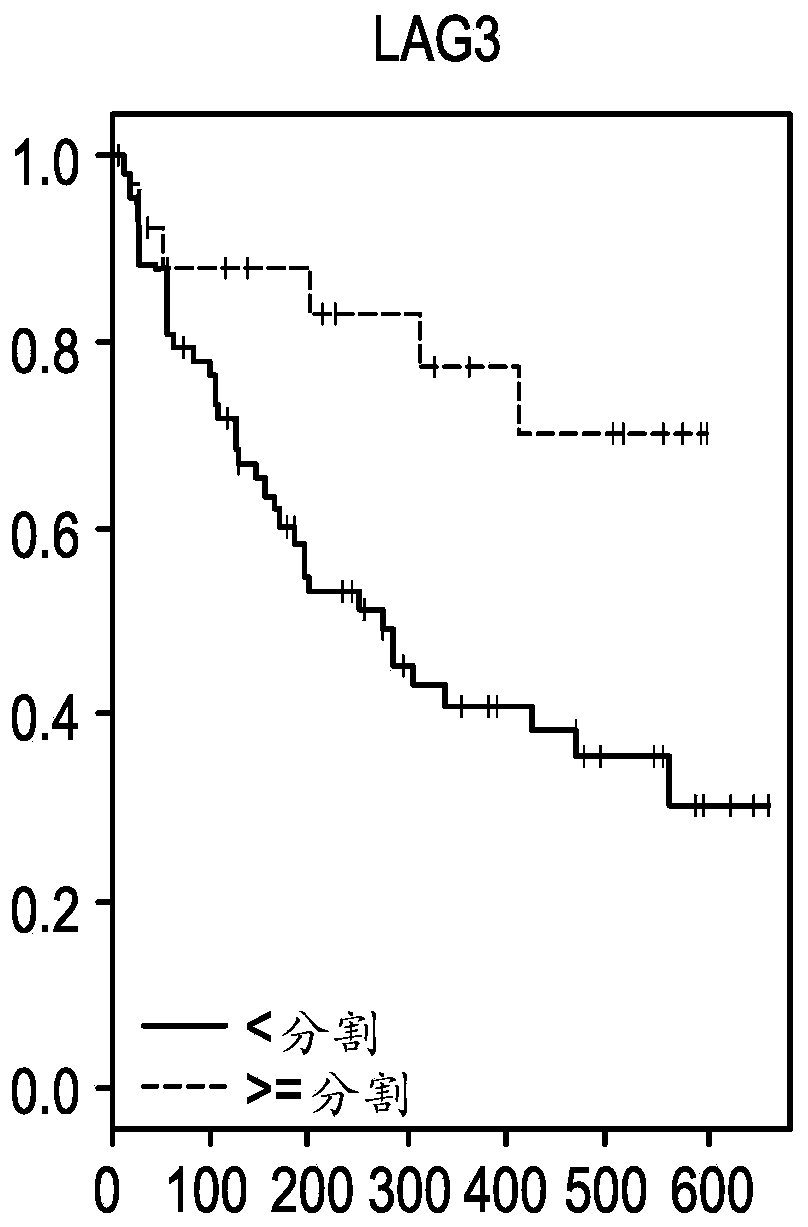
[0118] Example 2: CXCL9, CD274, LAG3 and IFNG are associated with overall response (OS) or progression-free response (PFS) in NSCLC
[0119] Patients were divided into high or low expression groups using the same cutpoints for each of the top four genes identified from the ORR analysis (CXCL9, CD274, LAG3, and IFNG). Kaplan-Meier estimates (KM) analyzes were then performed using overall response (OS) or progression-free response (PFS) as endpoints for each gene individually ( Figures 1A-1H ). Figures 1A-1D Panels showing overall responses to CXCL9, IFNG, LAG3 and CD274 (PDL1 ) in NSCLC patients. Each panel was segmented into high or low expression using cutpoints identified by ROC. Figures 1E-1H Panels showing progression-free survival for CXCL9, IFNG, LAG3 and CD274 (PDL1) in NSCLC patients. Each panel was segmented into high or low expression using cutpoints identified by ROC.
[0120] Using the log-rank test, all four genes showed statistical differences between the h...
example 3
[0121] Example 3: CD274(PDL1) did not show significant induction after treatment
[0122] Significant induction of each of the four genes (CXCL9, CD274, LAG3 and IFNG) following treatment with durvalumab was assessed. Paired t-tests were calculated between post-treatment and pre-treatment time points for each gene individually. The results are reported in Table 3. Only CD274 (PDL1 ) showed no significant induction at α=0.05 after treatment. These results indicated that three of the four genes were induced in tumors by durvalumab, suggesting immune activation of relevant immune-specific genes caused by treatment with this molecule.
[0123] Table 3. The therapeutic effect of durvalumab on four genes in NSCLC
[0124]
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com