A srap molecular marker primer combination and analysis method for genetic diversity analysis of P.
A technology of genetic diversity and molecular markers, applied in the field of SRAP molecular marker primer combination and analysis, to achieve high polymorphism, strong specificity, and uniform distribution
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0083] Of the 33 samples of Chonglou used in the present invention, 32 were collected from 6 areas in Jinzhai County, Anhui Province, and 1 was collected from Mount Huangshan. See Table 1 for details.
[0084] Table 1 Test material number and sampling point information
[0085]
[0086]
Embodiment 2
[0088] Extraction of Genomic DNA of Chonglou
[0089] The present invention collects the young leaves of Chonglou in Example 1 for the extraction of whole genome DNA, uses the CTAB method to extract the total genomic DNA of Chonglou, and dissolves the extracted DNA sample in TE buffer and places it at -20°C for later use. Before amplification, the DNA sample was diluted to 10 ng / μl with double distilled water, which was used as a template for PCR amplification reaction.
[0090] PCR reaction system and primers for SRAP marker
[0091] After seven pairs of primers, me2-em10, me3-em5, me3-em7, me3-em9, me3-em10, me4-em1 and me4-em4, each pair of primers was tested three times, the SRAP-PCR 25μl reaction system 10x PCR buffer (without Mg2+) 3.0μl, Mg2+1.5mmol / L, template DNA 40ng, dNTPs 0.25mmol / L, primer 0.4μmol / L, Taq enzyme 1U, the comprehensive amplification effect is the best. The PCR amplification reaction program was: 94°C pre-denaturation for 5 min, 94°C denaturation fo...
Embodiment 3
[0100] results and analysis
[0101] (1) Polymorphism analysis
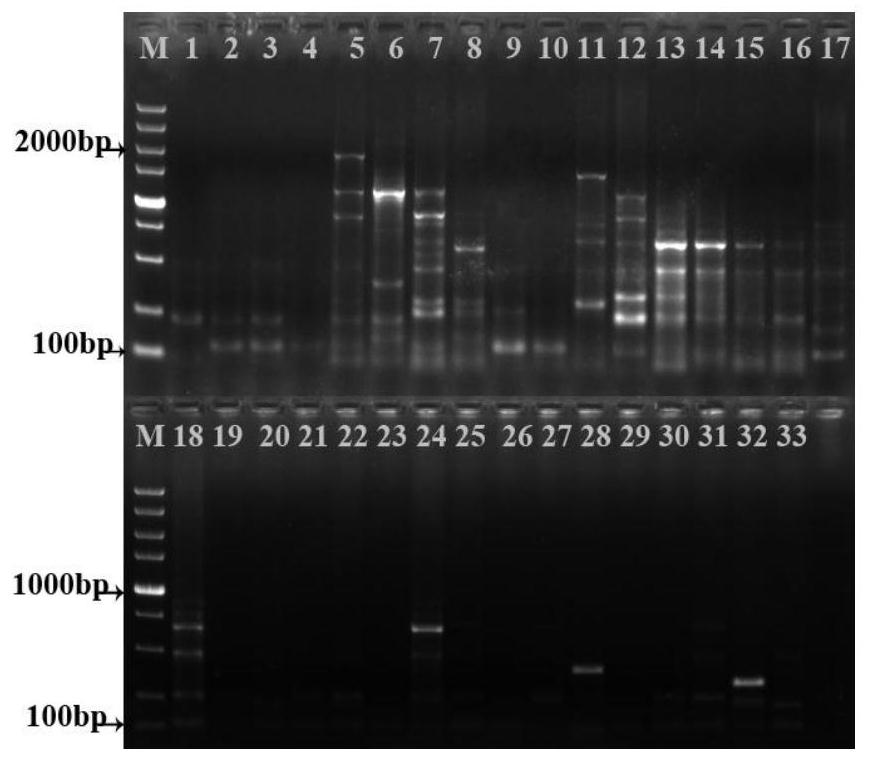
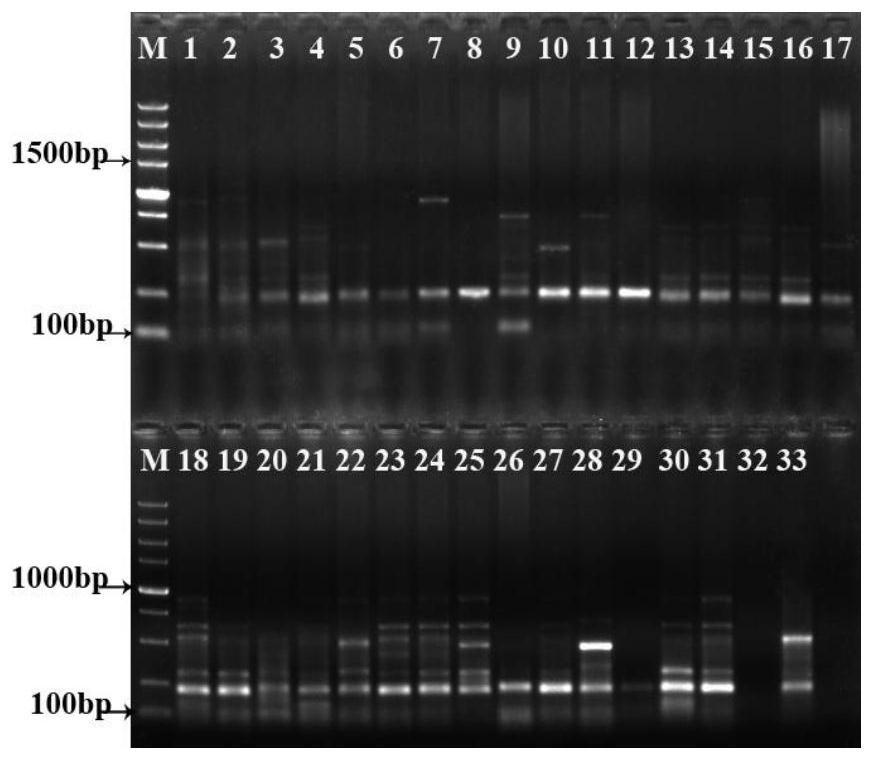
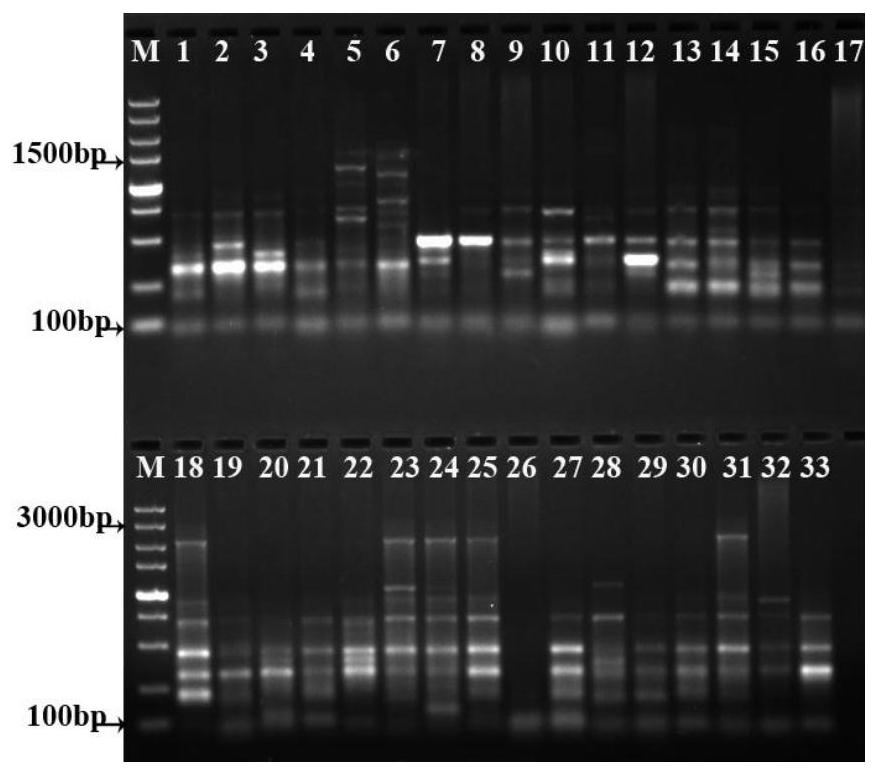
[0102] Figure 1-7 It is the electrophoresis diagram of the amplification products of 33 samples of double building with 7 pairs of primer combinations. It can be seen from the figure that the obtained bands are clear and abundant. The primer amplification results are shown in Table 3. A total of 101 bands were amplified by 7 pairs of primer combinations, including 97 polymorphic bands, and the polymorphic percentage (PPB) was 96.04%. The bands amplified by different primers are different, and the bands amplified by the same primers and different regions are also different, indicating that there are abundant polymorphisms and complex genetic backgrounds among the 33 samples. Relationships offer possibilities.
[0103] Table 3 Primer amplification results
[0104]
[0105]
[0106] Note: TNP: total number of amplified bands; NPB: number of polymorphic bands; PPB: percentage of polymorphism; PIC: informat...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com