Yeast expressing a synthetic calvin cycle
A Calvin cycle, yeast technology, applied in biosynthesis, fermentation, enzymes, etc.
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0442] Embodiment 1 prepares culture medium
[0443] LB medium is used for the cultivation of Escherichia coli DH 10B, and the operation steps are as follows.
[0444] Prepare LB medium (10.0g*L -1 Soy peptone (Quest), 5.0g*L -1 Yeast extract (MERCK) and 5.0g*L -1 NaCl, and adjusted the pH value to 7.4-7.6 with 4N NaOH), and distributed in 500mL Schott bottles. The sterilization method is autoclaving at 121°C for 20 minutes.
[0445] Yeast peptone (YP) medium was used to cultivate Pichia pastoris CBS7435 wt in shake flasks, and the operation steps were as follows.
[0446] Before adding carbon source, YP-medium (20.0g*L -1 Soy peptone (Quest), 10.0g*L -1 Yeast extract (MERCK, pH adjusted to 7.4–7.6 with 4N NaOH) was autoclaved. Prepare 10 times glucose stock solution (220g*L -1 D(+)-glucose monohydrate) and sterilized by autoclaving. Add 10 times the glucose stock solution to the YP medium at a ratio of 1 / 10 to obtain a YPD medium.
[0447] Glycerol-containing batch ...
Embodiment 2
[0459] Example 2 Constructing plasmids and linear DNA fragments
[0460] All expression cassettes (promoter, CDS, terminator) were constructed by Golden Gate cloning (Engler et al.PloSOne 4(5):e5553.doi:10.1371 / journal.pone.0005553) and flanked by the corresponding integration sites point to replace the above three genes, namely aox1, das1 and das2. Follow the plasmid DNA construct operating procedure published in (Sarkari, et al.2017BioresourceTechnology.doi:10.1016 / j.biortech.2017.05.004) to realize the operation for constructing all linear DNA fragments and guide RNA (gRNA) / hCas9 plasmid process.
[0461] The coding sequences (CDS) of the genes described in Table 5 were combined with the methanol-inducible promoter and terminator sequences of Pichia pastoris CBS7435 wt (Table 6).
[0462] Table 5: Creation of the synthetic Calvin cycle in Pichia pastoris (Centraalbureau voorSchimmelcultures, NL, genome sequenced by (Küberl et al., 2011; Valli et al., 2016)) according to e...
Embodiment 3
[0476] Example 3 Construction of a Pichia pastoris strain expressing a functional Calvin cycle targeting peroxisomes
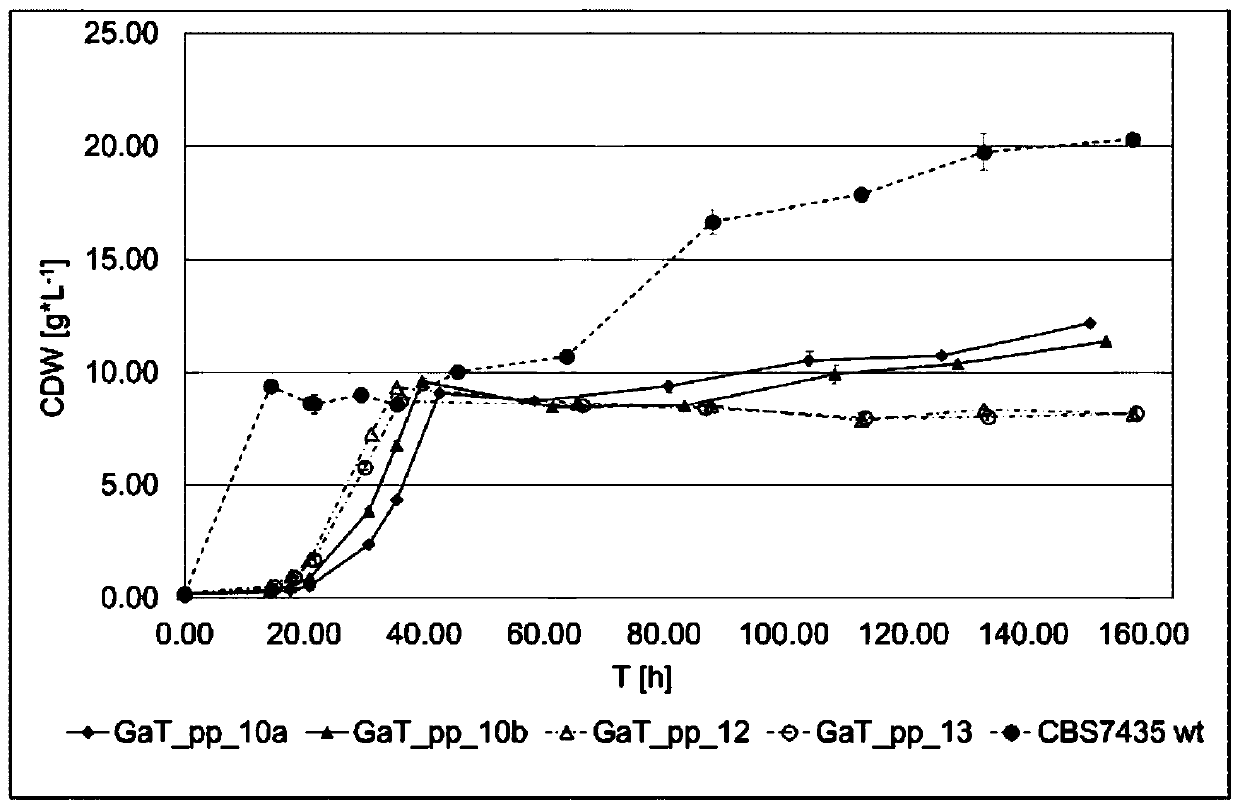
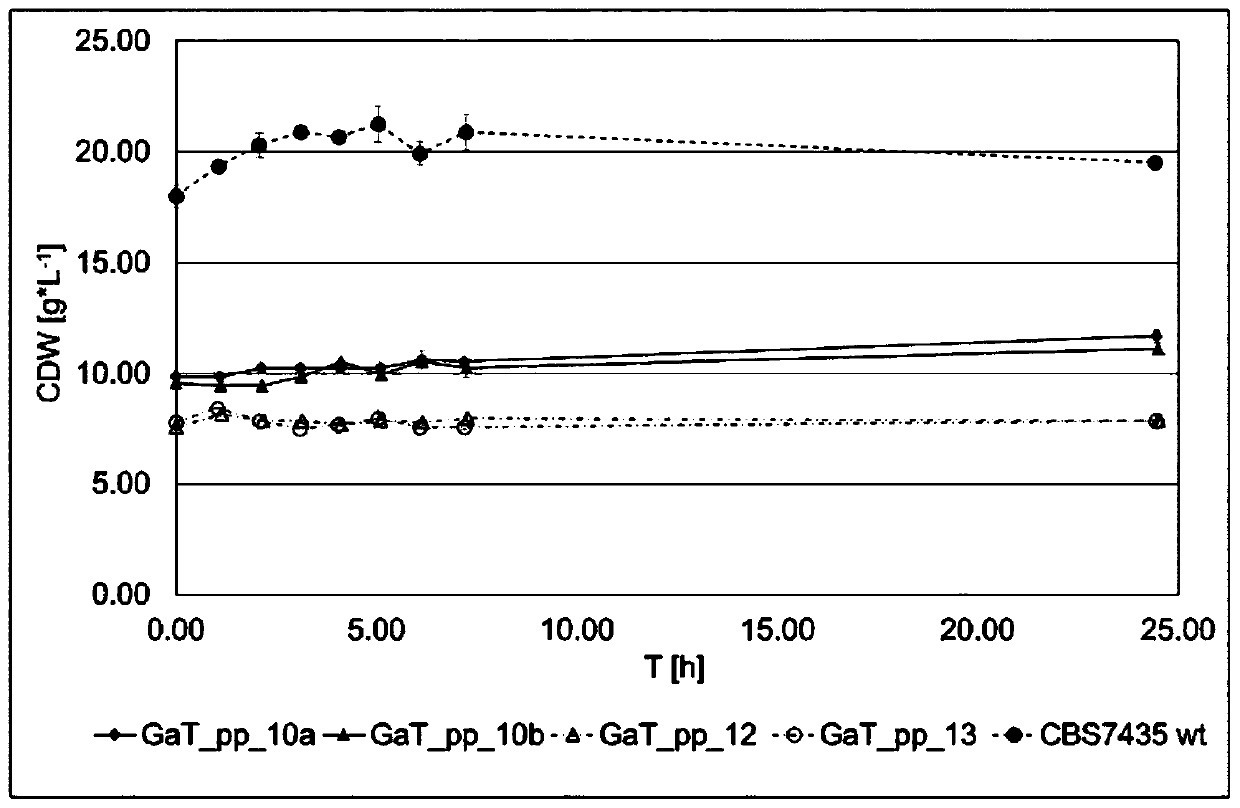
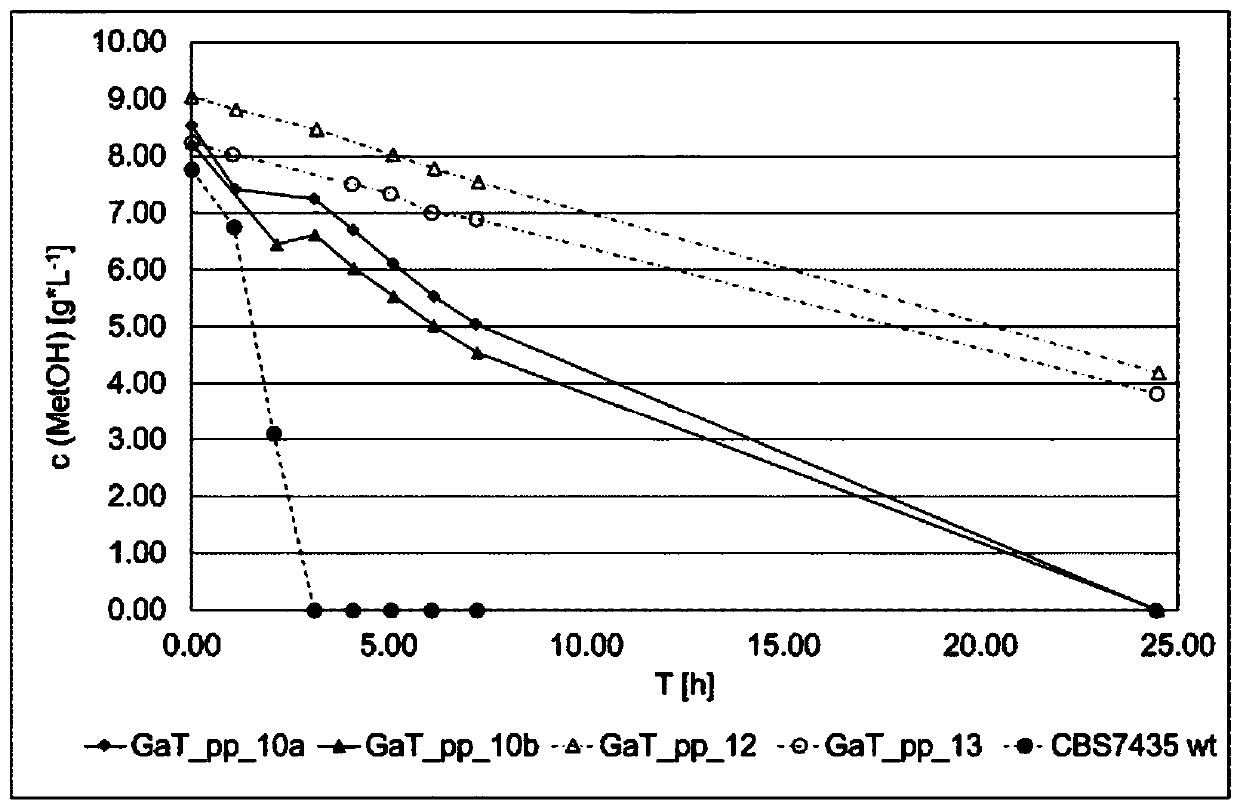
[0477] To create GaT_pp_10 and control Pichia pastoris strains, three genes in the Pichia pastoris genome were deleted, namely AOX1 (ORF ID: PP7435_Chr4-0130), DAS1 (ORF ID: PP7435_Chr3-0352) and DAS2 ( ORF ID: PP7435_Chr3-0350), and 8 genes PGK1, TDH3, TPI1, PRK, TKL, GroEL, GroES and RuBisCO (Table 5 and Table 6) were integrated into the genome.
[0478] 3.1 Transformation of Pichia pastoris
[0479] Pichia pastoris transformation was performed on chemically competent cells by electroporation as described below. A single colony of Pichia pastoris (CBS7435 wt or the corresponding clone) was inoculated into 10 mL of YPD pre-culture, and cultured overnight ( o ver n ight) (o / n; ~16h) (shaker; 180rpm; 28°C). The next day, 100 mL of the main culture was inoculated. The inoculum volume of the preculture was calculated as follows so that the main culture reach...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com