CRISPR off-target effect prediction method based on deep learning
An off-target effect and deep learning technology, applied in the field of bioinformatics, can solve problems such as low efficiency and inability to effectively use genome prior information, and achieve the effect of improving accuracy
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment Construction
[0048] The present invention will be further described below in conjunction with the accompanying drawings and embodiments.
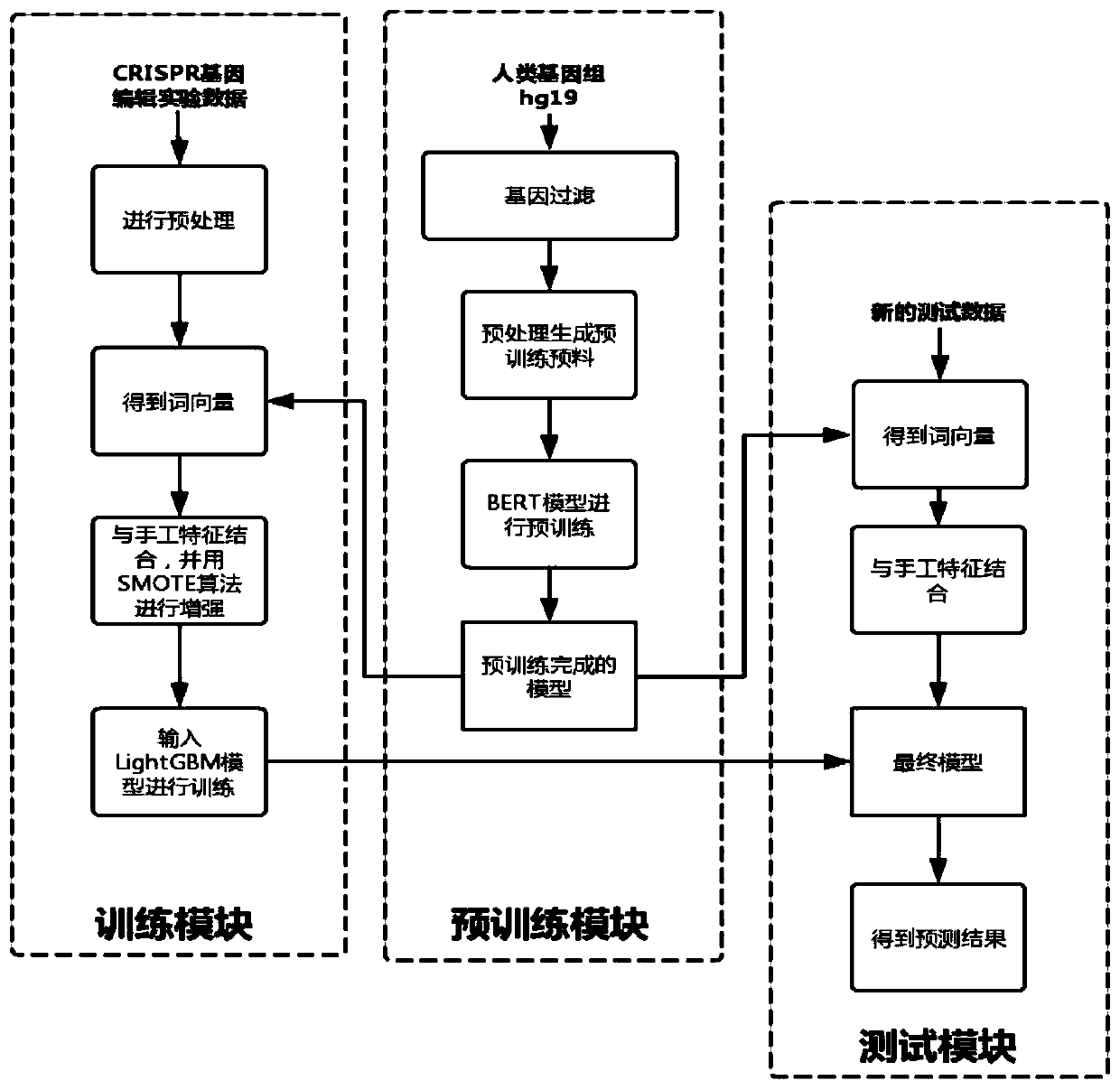
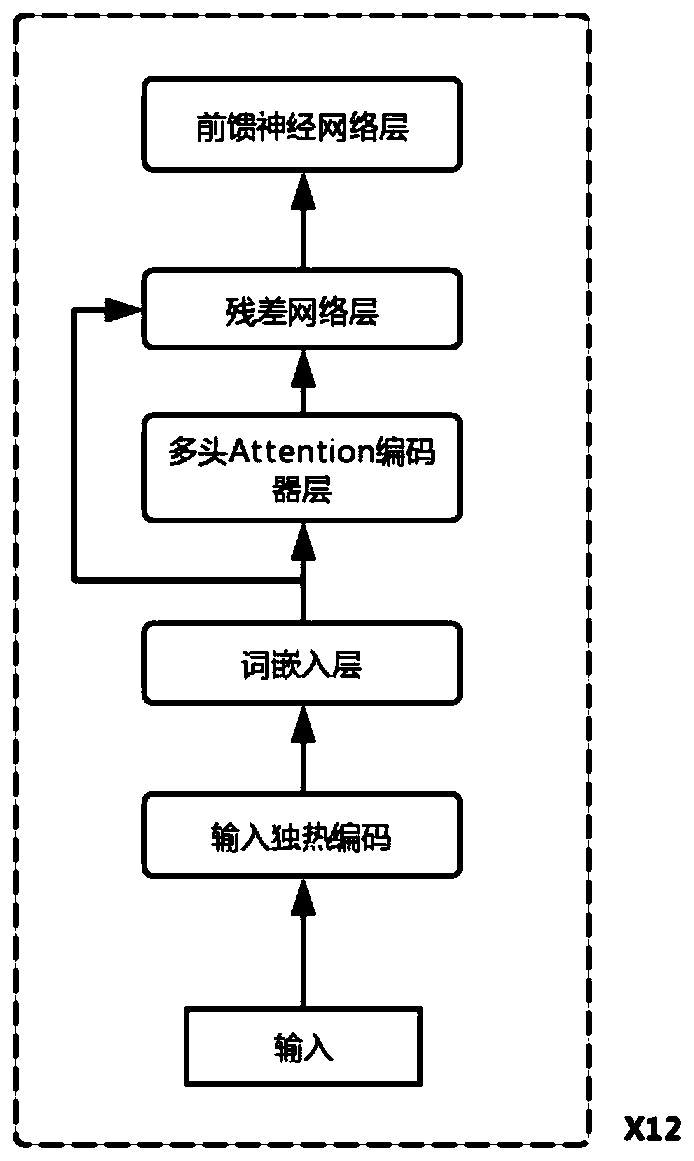
[0049] like figure 1 As shown, the steps of the method are described in detail below in conjunction with an example:
[0050] A method for predicting off-target effects of CRISPR based on deep learning, comprising the following steps:
[0051] The first step is to start filtering all the genes of the human genome hg19: In order to avoid the reduction of subsequent prediction accuracy caused by too large pre-trained data, it is necessary to filter out the data irrelevant to the task first, and find out all PAM sequences as NGG in a targeted manner , and then predict the sequence pair whose PAM sequence is NGG;
[0052] The second step is to preprocess the original corpus, which is divided into the following four steps:
[0053] 1. First split the sequence with spaces as intervals;
[0054] 2. Construct sequence sample pairs and randomly combine the two...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com